Are you looking for an essay on the ‘Transcription and Its Types’? Find paragraphs, long and short essays on the ‘Transcription and Its Types’ especially written for school and college students.
Essay # 1. Introduction to Transcription:
In transcription, a section of DNA (a gene), carrying the genetic code for the synthesis of a specific protein molecule, is copied into mRNA. Messenger RNA then migrates to the cytoplasm where ribosomes are then translating the code to construct the protein.
Transcription has similarities to DNA replication but only involves a small portion of the DNA molecule. In transcription, the DNA will unzip between the nitrogenous bases and expose the sequence of a gene that codes for a specific protein. RNA nucleotides, present in the nucleus, will then move in and complementary base pair with one side of the unzipped DNA. The complementary base pairing is the same with the exception of Uracil which replaces Thymine (e.g. an adenine nucleotide in DNA will transcribe a uracil nucleotide in the mRNA).
The RNA nucleotides are joined together by an enzyme called RNA polymerase. The mRNA then moves out of the nucleus through nuclear pores to deliver the coded message to the ribosomes in the cytoplasm. Transcription also produces ribosomal RNA (rRNA) and transfer RNA (tRNA) that are used in protein synthesis. rRNA is a component of ribosomes and tRNA delivers amino acids to the ribosome during translation.
DNA sequence is enzymatically copied by RNA polymerase, RNA polymerase binds to a special region called promoter region. This the start point surrounds the first base pair that is transcribed into RNA, from this point RNA polymerase moves along the template, synthesizing RNA until it reaches a terminator sequence. This action defines a complete transcription unit that extends from the promoter to the terminator.
The stretch of DNA that is transcribed into an RNA molecule is called a transcription unit. A DNA transcription unit that is translated into protein contains sequences that direct and regulate protein synthesis (Fig. 8.3). In addition to the coding sequence that is translated into protein, there is a regulatory sequence located before (towards the 5′ DNA end) the coding sequence, it is called 5′ untranslated region (5’UTR) and sequence found towards the 3′ DNA end.
RNA Polymerases:
RNA Polymerases are DNA-directed RNA polymerase. They are essential to life and are found in all organisms and many viruses. In chemical terms, they are nucleotidyl transferase that polymerizes ribonucleotides at the 3′ end of an RNA transcript.
(a) Prokaryotic RNA Polymerase:
E. coli has a single DNA-directed RNA polymerase that synthesizes all types of RNA. It is a large and complex enzyme, containing five core subunits and a sixth subunit, called Sigma factor (σ) that binds transiently to the core enzyme and directs the enzyme to for initiation sites on the DNA (Sigma factor (σ) binds to the initiation site). These six subunits constitute the RNA polymerase holoenzyme (Holoenzyme = Core enzyme + Sigma factor).
The σ sub- unit is weakly bound and can be separated from the other subunits, yielding a core polymerase consisting of two α, one β, one β’ and one 00 subunits (α2ββω; MW. 390,000) (Table 8.1). The core polymerase is fully capable of catalyzing the polymerization of NTPs into RNA, indicating that o is not required for the basic catalytic activity of the enzyme. The core polymerase does not bind specifically to the DNA sequences that signal the normal initiation of transcription; therefore, the σ subunit is required to identify the correct sites for initiation of transcription. Sigma factor will be released after initiation (Fig. 8.4).
RNA polymerase requires a DNA template, ATP, GTP, UTP, and CTP as precursors of the nucleotide units of RNA, as well as Mg2+. The purified enzyme also contains Zn2+. RNA polymerase elongates an RNA strand by adding ribonucleotide units to the 3′-hydroxy 1 end of the RNA chain and thus builds RNA chains in the 5’—>3′ direction. RNA synthesis takes place within a “transcription bubble,” in which DNA strands are transiently separated into its single strands and the template strand is used to direct synthesis of the RNA strand.
(b) Eukaryotic RNA Polymerase:
There are three classes of eukaryotic RNA polymerases:
I, II and III, each comprising of two large subunits and 12-15 smaller subunits. The two large subunits are homologous to the E. coli β and β’ subunits. Two smaller subunits are similar to the E. coli α sub- unit. However, the eukaryotic RNA polymerase does not contain any subunit similar to the E. coli Sigma factor. Therefore, in eukaryotes, transcriptional initiation should be mediated by other proteins (Table 8.2).
* The three RNAPs are distinguished on the basis of their sensitivity towards transcription inhibitors like α – amanitin.
Promoters:
A promoter is a regulatory region of DNA located upstream (towards the 5′ region) of a gene, providing a control point for regulated gene transcription. The promoter contains specific DNA sequences that are recognized by proteins known as transcription factors. These factors bind to the promoter sequences, recruiting RNA polymerase, the enzyme that synthesizes the RNA from the coding region of the gene (Fig. 8.5).
Enhancers:
Enhancers act to stimulate the activity of certain promoters. In common with upstream promoter elements they can be active in all tissues or can display tissue-specificity. They are modular in structure and can act co-operatively. Many enhancers seem to contain multiple binding sites for transcription factors which interact and often these are sites which are also found in promoter sequences. Enhancers are thought to act by binding transcription factors, to form an enhanceosome (a protein complex that binds to the “enhancer” region of a gene, found upstream or downstream, of the promoter, or within a gene) and looping out the DNA between the enhancer and the promoter thus bringing factor binding sites together, increasing efficiency of recruitment of transcription factors to the promoter and therefore transcription initiation.
Essay # 2. Types of Transcription:
A. Prokaryotic Transcription Process:
Much of the pioneering work on transcription was carried out in prokaryotes, most notably in the bacterium E. coli. These studies laid the foundation for work that was later carried out in the more complex eukaryotes.
Transcription can be divided into three phases:
i. Initiation,
ii. Elongation, and
iii. Termination.
i. Initiation:
Initiation describes the formation of the first nucleotide bonds in RNA. It starts with template recognition which begins with the binding of RNA polymerase holoenzyme to the double-stranded DNA at a promoter to form a “closed complex”. Then the strands of DNA are separated to form the “open complex” that makes the template strand available for base pairing with ribonucleosides. The transcription bubble is created by localized unwinding that begins at the site where RNA polymerase binds. RNA polymerase binds to the promoter in at least two distinguishable steps. The holoenzyme first binds the DNA and migrates to the – 35 region, forming the “closed complex.”
The DNA is then unwound for about 17 base pairs beginning at the -10 region, exposing the template strand at the initiation site. The RNA polymerase binds more tightly to this unwound region; forming an “open complex” for RNA synthesis, to begin. The initiation phase ends when the enzyme succeeds in extending the chain and clears the promoter. The sigma factor eventually dissociates from the holoenzyme after recognizing the initiation site and elongation proceeds.
Promoters can differ in “strength,” or how actively they promote transcription of their adjacent DNA sequence. Promoter strength is in many (but not all) cases, a matter of how tightly RNA polymerase and its associated accessory proteins bind to their respective DNA sequences (Fig. 8.8). Most transcripts originate using adenosine-5′-triphosphate (ATP) and, to a lesser extent, guanosine -5′-triphosphate (GTP) (purine nucleoside triphosphates) at the +1 site.
ii. Elongation:
Elongation is the function of the RNA polymerase core enzyme. RNA polymerase moves along the template, locally “unzipping” the DNA double helix. This allows a transient base pairing between the incoming nucleotide and newly-synthesized RNA and the DNA template strand. As it is made, the RNA transcript forms secondary structure through intra-strand base pairing. The average speed of transcription is about 40 nucleotides per second, much slower than DNA polymerase. Other protein factors may bind to polymerase and alter the rate of transcription and some specific sequences are transcribed more slowly than others. Eventually, RNA polymerase comes to the end of the region under transcription.
iii. Termination:
Termination depends on the protein factor. Rho factor is a protein involved in assisting E. coli RNA polymerase to terminate transcription at certain terminators (called rho-dependent terminators). Rho-dependent terminators are sequences that terminate transcription by bacterial RNA polymerase in the presence of the rho factor.
Rho-independent terminators have a characteristic structure, which features a strong G-C rich stem and loop, a sequence of 4-6 U residues in the RNA, which are transcribed from a corresponding stretch of A in the template. Intrinsic terminators are able to terminate transcription by bacterial RNA polymerase in the absence of any additional factors. Core enzyme can terminate in vitro at certain sites in the absence of any other factor.
There are ~1100 sequences in the E. coli genome that fit these criteria, suggesting that about half of the genes have intrinsic terminators. All hairpins that form in the RNA product cause the polymerase to slow (and perhaps to pause) in RNA synthesis. Pausing occurs at sites that resemble terminators but have an increased separation (typically 10-11 bases) between the hairpin and the U-run. If the pause site does not correspond to a terminator, usually the enzyme moves on again to continue transcription. The length of the pause varies, but at a typical terminator lasts -60S.
B. Eukaryotic Transcription Process:
Most of the eukaryotic protein-coding genes contain segments called introns, which break up the amino acid coding sequence into segments called exons. The transcript of these genes is resulted as pre-mRNA (precursor-mRNA). The pre-mRNA is then processed in the nucleus where the introns are removed and the exons are spliced together into a translatable mRNA. The mRNA subsequently released from the nucleus and are translated in to proteins in the cytoplasm by ribosomes (Fig. 8.9).
RNA Polymerase:
Although transcription in eukaryotes is similar to that in prokaryotes, the process appears to be complex. Instead of one RNA polymerase, there are three (RNA polymerases I, II, and III) involved in eukaryotic transcription. RNA polymerase I (localized to the nucleolus) transcribes the rRNA precursor molecules. RNA polymerase II produces most mRNAs and snRNAs. RNA polymerase III is responsible for the production of pre-tRNAs, 5SrRNA and other small RNAs.
The mitochondria and chloroplasts have their own RNA polymerases. There is considerable relatedness between the three eukaryotic RNA polymerases and to the prokaryotic E. coli RNA polymerase, especially between the largest and second largest sub- units. RNA polymerase II differs from the others in that the largest subunit has a carboxy terminal extension called the carboxy terminal domain (CTD). The CTD contains a highly repeated heptapeptide Tyr-Ser-Pro-Thr-Ser-Pro-Ser, which can be heavily phosphorylated. This phosphorylated domain is essential for transcription by RNA polymerase II in most eukaryotes, and it also links the processes of transcription and RNA processing.
(a) RNA Polymerase I:
RNA polymerase I is a complex of 13 subunits. Promoters for RNA polymerase I contain two important regulatory elements, a core promoter located from around 20 nucleotides downstream of the transcription initiation site to around 40 nucleotides upstream, and an upstream control element (UCE) situated some 100 nucleotides upstream of the transcription initiation site. The first step in formation of the polymerase I transcription initiation complex is binding of two molecules of Upstream Binding Factor (UBF), one to the UCE and the other to the core promoter element. Interaction between the two UBFs bound to the promoter result in looping out of the intervening promoter DNA and provision of a promoter structure that can be recognized, and bound by, in humans, the core promoter element- binding factor, selectivity factor 1 (SL1). SL1 is a multi-subunit protein composed of TATA -binding protein (TBP) and three TBP-associated factors to which RNA polymerase I binds to complete the transcription initiation complex.
(b) RNA Polymerase II:
Formation of the stable transcription initiation complex on the promoter of a gene is directed by a DNA sequence with the consensus TATA/TAA/T, around 30 nucleotides upstream of the transcription initiation site of the gene, the so-called TATA box. Mutations in the TATA box sequence have a striking effect on transcription; few mutations are tolerated, demonstrating that this is a crucial sequence. The primary transcription complex, TFIID, consisting of up to 12 different TAFs, forms on TBP bound to the TATA box, and acts to direct RNA polymerase II to the correct transcription initiation site that serves as a focus for formation of the stable transcription initiation complex. A huge multi-protein structure is thus assembled in a highly ordered manner on TFIID.
(c) RNA Polymerase III:
The promoters for genes transcribed by RNA polymerase III are located downstream of the transcription initiation site, within the coding region of the gene. For tRNA genes a large multi-subunit protein, TFIIIC, binds with high affinity to the so-called “B-box” within the gene, and with lower affinity to the “A-box” upstream. TFIIIC acts as an assembly site for recruitment of the TFIIIB trimeric complex (containing TBP), which has no distinct sequence requirement for binding. The TFIIIB/C complex is capable of binding RNA polymerase III and initiating transcription. For 5S RNA there is only one promoter box within the gene, the “C-box”, which binds TFIIIA. This factor binds TFIIIC, as for the tRNA genes and positions it at a similar distance with respect to the transcription initiation site. TFIIIB can then bind followed by RNA polymerase III as before.
The Promoter:
Eukaryotic nuclear genes have three classes of promoters which are specific for the three types of RNA polymerases.
The promoter for RNA polymerase I has two components:
(1) A core promoter (surrounding the start point) and
(2) An upstream control element.
The core promoter region is located from -31 to +6 around the transcription start point. Another sequence further upstream, called the upstream control element (UCE), located from -187 to -107 is also required for efficient transcription. Both elements are closely related; there is approximately 85% sequence identity between them. These elements are also unusual in that they are GC-rich. In general, sequences around the start-point of transcription tend to be AT-rich so that melting of the DNA duplex is easier.
After the binding of appropriate transcription factors to both sites, RNA polymerase I binds to the core promoter. The typical promoter for RNA polymerase II has a short initiator sequence, consisting mostly of pyrimidines and usually a TATA box about 25 bases upstream from the start point. This type of promoter (with or without the TATA box) is often called a polymerase II core promoter, because for most genes a variety of upstream control elements also play important role in the initiation of transcription. The promoters for RNA polymerase III vary in structure but the ones for tRNA genes and 5S rRNA genes are located entirely downstream of the start point, within the transcribed sequence.
General transcription factors and the polymerase undergo a pattern of sequential binding to initiate transcription of nuclear genes. TFIID binds to the TATA box followed by the binding of TFIIA and TFIIB. The resulting complex is now bound by the polymerase, to which TFIIF has already attached. The initiation complex is completed by the addition of TFIIE, TFIIJ, and TFIIH. After an activation step requiring ATP-dependent phosphorylation of the RNA polymerase molecule, the polymerase can initiate transcription at the start point (Fig. 8.10).
The TATA-binding protein (TBP) is a subunit of the TFIID and plays a role in the activity of both RNA polymerase I and RNA polymerase III transcription. TBP is also essential for transcription of TATA-less genes. TBP differs from most DNA-binding proteins in that it interacts with the minor groove of DNA, rather than the major groove and imparts a sharp bend to the DNA. The TBP has been highly conserved during evolution. When TBP is bound to DNA, other transcription-factor proteins can interact with the convex surface of the TBP saddle. TBP is required for transcription initiation on all types of eukaryotic promoters.
Termination signals end the transcription of RNA by RNA polymerase I and RNA polymerase III without the activity of hairpin structures as seen in prokaryotes. mRNA is cleaved 10 to 35 base-pairs downstream of a AAUAAA sequence (which acts as a poly-A tail addition signal). Ribosomal RNA processing involves cleavage of multiple rRNAs from a common precursor. The eukaryotic transcription unit that includes the genes for the three largest rRNAs occurs in multiple copies and arranged in tandem arrays with non -transcribed spacers separate the units.
Each transcription unit includes the genes for the three rRNAs and transcribed spacer regions. The transcription unit is transcribed by RNA polymerase I into a single long transcript (pre-rRNA) with a sedimentation coefficient of about 45S. RNA processing yields mature rRNA molecules (Fig. 8.11). RNA cleavage actually occurs in a series of steps which varies in order with the species and cell type but the final products are always the same three types of rRNA molecules.
Every tRNA gene is transcribed as a precursor that must be processed into a mature tRNA molecule by the removal, addition and chemical modification of nucleotides. Processing for some tRNA involves removal of the leader sequence at the 5′ end, replacement of two nucleotides at the 3′ end by the sequence CCA (with which all mature tRNA molecules terminate), chemical modification of certain bases and excision of an intron. The mature tRNA is often diagrammed as a flattened cloverleaf which clearly shows the base pairing between self-complementary stretches in the molecule.
Messenger RNA in eukaryotes is first made as heterogeneous nuclear mRNA (or pre- mRNA) then processed into mature mRNA through the addition of a 5′ cap structure, addition of poly-A tails and the splicing out of introns. To give the mRNA stability, a 5′ “cap” (a guanosine nucleotide methylated at the 7th position) is joined to the first nucleotide in an unusual 5’ to 5’ linkage (sort of “backwards”). During the capping process, the first two nucleotides of the message may also become methylated.
Transcription of eukaryotic pre-mRNAs often proceeds beyond the 3′ end of the mature mRNA. An AAUAAA sequence located slightly upstream from the proper 3′ end then signals that the RNA chain should be cleaved about 10—35 nucleotides downstream from the signal site, followed by addition of a poly-A tail catalyzed by poly(A) polymerase.
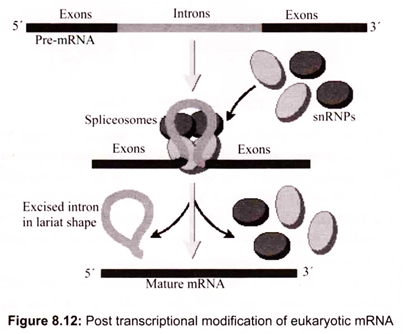
Spliceosomes remove introns from pre-mRNA. Introns were discovered to exist in eukaryotic mRNA by mixing mature mRNA molecules with the genes (DNA) from which they had been transcribed and examining the hydrogen bonded hybrids under an electron microscope. Hybridization of a eukaryotic mRNA molecule with a gene which has one intron will produce two single-stranded DNA loops where the mRNA has hybridized to the DNA template strand plus an obvious double-stranded DNA loop. The double-stranded DNA loop represents the intron, which contains sequences that do not appear in the final mRNA. Restriction enzyme analysis has revealed the presence of introns (Fig. 8.12).
The spliceosome is an RNA-protein complex that splices intron-containing pre-mRNA in the eukaryotic nucleus. The substrate here is a molecule of pre-mRNA with two exons and one intron. In a stepwise fashion, the pre-mRNA assembles with the U1 snRNP, U2 snRNP, and U4/U6 and U5 snRNPs (along with some non-snRNP splicing factors), forming a mature spliceosome.
The pre-mRNA is cleaved at the 5′ splice site and the newly released 5′ end is linked to an adenine (A) nucleotide located at the branch-point sequence, creating a looped lariat structure. Next the 3′ splice site is cleaved and the two ends of the exon are joined together, releasing the intron for subsequent degradation. Alternative splicing may results in the formation of alternate forms of mRNA. Most mRNA molecules have a high turnover rate as the molecules are rapidly degraded and replaced. tRNA and rRNAs are relatively stable. Bacterial mRNAs have half-lives of a few minutes, while eukaryotic mRNA range from hours to days. Transcription allows amplification of the genetic information because many copies of the mRNA can be produced to direct a great deal of protein synthesis.
TATA Binding Protein (TBP):
TATA binding protein (TBP) is the general transcription factor common to all 3 RNA polymerase complexes. TBP is a 38 kDa saddle-shaped monomer which can contact and bend severely TATA-containing DNA in the minor groove. TBP therefore changes the conformation of the DNA and this is thought to facilitate transcription factor binding. It is highly conserved from yeast to mammals and is evolutionarily ancient, a related protein being found in archebacteria. TBP presents a wide outer surface for simultaneous binding of a number of TBP-associated factors (TAFs) and the complexes these form are the positioning factors for RNA polymerase. TAFs appear to regulate the activity of TBP and together they determine the specificity of polymerase binding to promoters.
Types of Promoters Used to Regulate Gene Expression:
Promoters used in biotechnology are of different types, according to the intended type of control of gene expression. Eukaryotic promoters are usually larger than just the TATA box and initiation site. They tend to be modular in architecture and can be very complex, if they are regulated in response to tissue type, differentiation stage, or cellular signals.
Other transcription factor binding sites are generally found upstream of the TATA box and act through binding of different transcriptional activators and/or repressors to increase or decrease the level of transcription (i.e. increase or decrease the rate of loading of the RNA polymerase complex on the promoter).
They can be generally divided into:
a. Constitutive Promoters:
Constitutive factors interact with the basal transcription initiation complex to increase levels of transcription in all tissues. In the absence of these factors the basal promoter can provide only low levels of transcription initiation and binding of one or more of these factors is necessary for significant levels of transcription to occur. All these elements are essential for full transcriptional activity in promoters where they are present.
The CCAAT box, at least, may be recognized by a variety of proteins that can activate or repress gene expression. These promoters direct expression in virtually all tissues and are largely, if not entirely, independent of environmental and developmental factors. As their expression is normally not conditioned by endogenous factors, constitutive promoters are usually active across species and even across kingdoms.
b. Tissue-Specific or Development-Stage-Specific Promoters:
These direct the expression of a gene in specific tissue(s) or at certain stages of development. For plants, promoter elements that are expressed or affect the expression of genes in the vascular system, photosynthetic tissues, tubers, roots and other vegetative organs, or seeds and other reproductive organs can be found in heterologous systems (e.g. distantly related species or even other kingdoms) but the most specificity is generally achieved with homologous promoters (i.e. from the same species, genus or family). This is probably because the coordinate expression of transcription factors is necessary for regulation of the promoter’s activity.
c. Inducible Promoters:
Inducible factors are produced in response to specific cellular signals, e.g. stress, growth stimulation, metabolite concentrations etc. They interact with their appropriate promoter binding sites to modulate gene expression. Some examples are hormone receptors, metal ions, heat shock proteins, and cAMP. The performance inducible factors are not conditioned to endogenous factors but to environmental conditions and external stimuli that can be artificially controlled.
Within this group, there are promoters modulated by abiotic factors such as light, oxygen levels, heat, cold and wounding. Since some of these factors are difficult to control outside an experimental setting, promoters that respond to chemical compounds, not found naturally in the organism of interest, are of particular interest. Along those lines, promoters that respond to antibiotics, copper, alcohol, steroids, and herbicides, among other compounds, have been adapted and refined to allow the induction of gene activity independent of other biotic or abiotic factors.
d. Synthetic Promoters:
Promoters are synthesized by bringing together the primary elements of a promoter region from diverse origins in a molecule.