Read this article to learn about the restriction enzymes and their mode of action. And it also describes different types of restriction enzymes. The types are: (1) Type I (2) Type II and (3) Type III.
Restriction enzymes are DNA-cutting enzymes found in bacteria (and harvested from them for use). Because they cut within the molecule, they are often called restriction endonucleases.
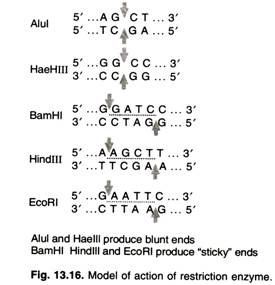
A restriction enzyme recognizes and cuts DNA only at a particular sequence of nucleotides. For example, the bacterium Hemophilus aegypticus produces an enzyme named Haelll that cuts DNA wherever it encounters the sequence
5’GGCC3’
3’CCGG5’
The cut is made between the adjacent G and C. This particular sequence occurs at 11 places in the circular DNA molecule of the virus phiX174. Thus treatment of this DNA with the enzyme produces 11 fragments, each with a precise length and nucleotide sequence. These fragments can be separated from one another and the sequence of each determined. Haelll and Alul cut straight across the double helix producing “blunt” ends. However, many restriction enzymes cut in an offset fashion.
The ends of the cut have an overhanging piece of single-stranded DNA. These are called “sticky ends” because they are able to form base pairs with any DNA molecule that contains the complementary sticky end (Fig. 13.16). Any other source of DNA treated with the same enzyme will produce such molecules. Mixed together, these molecules can join with each other by the base pairing between their sticky ends.
The union can be made permanent by another enzyme, DNA ligase that forms covalent bonds along the backbone of each strand. The result is a molecule of recombinant DNA (rDNA). Because recognition sequences and cleavage sites differ between restriction enzymes, the length and the exact sequence of a sticky-end “overhang”, as well as whether it is the 5′ end or the 3′ end strand that overhangs, depends on which enzyme produced it. Base-pairing between overhangs with complementary sequences enables two fragments to be joined or “spliced” by a DNA ligase.
A sticky-end fragment can be ligated not only to the fragment from which it was originally cleaved, but also to any other fragment with a compatible sticky end. The sticky end is also called a cohesive end or complementary end in some reference.
If a restriction enzyme has a non- degenerate palindromic (the sequence on one strand reads the same in the same direction on the complementary strand e.g. GTAATG is not a palindromic DNA sequence, but GTATAC is, GTATAC is complementary to CATATG) cleavage site, all ends that it produces are compatible. Ends produced by different enzymes may also be compatible.
Naming:
Restriction enzymes are named based on the bacteria in which they are isolated in the following manner:
E Escherichia (genus)
co coli (species)
R RY13 (strain)
I First identified Order ID’d in bacterium
Patterns of DNA Cutting by Restriction Enzymes:
(i) 5′ overhangs:
The enzyme cuts asymmetrically within the recognition site such that a short single-stranded segment extends from the 5′ ends. BamHI cuts in this manner.
(ii) 3′ overhangs:
Again, we see asymmetrical cutting within the recognition site, but the result is a single-stranded overhang from the two 3′ ends. Kpnl cuts in this manner.
(iii) Blunts:
Enzymes that cut at precisely opposite sites in the two strands of DNA generate blunt ends without overhangs. Smal is an example of an enzyme that generates blunt ends.
The 5′ or 3′ overhangs generated by enzymes that cut asymmetrically are called sticky ends or cohesive ends, because they will readily stick or anneal with their partner by base pairing.
Mode of Action:
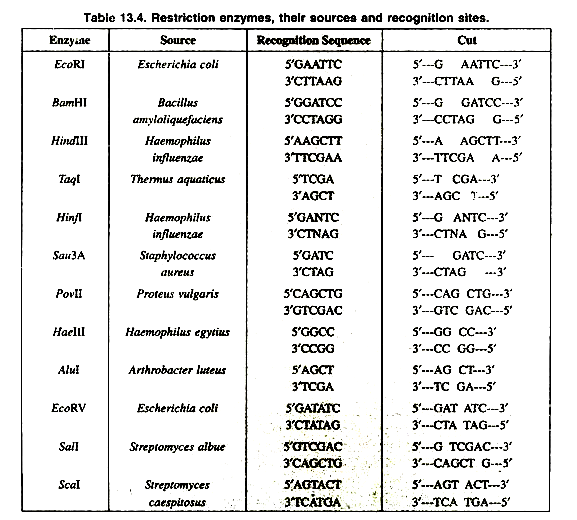
A restriction enzyme (or restriction endonuclease) is an enzyme that cuts double-stranded DNA (Table 13.4). The enzyme makes two incisions, one through each of the sugar-phosphate backbones (i.e., each strand) of the double helix without damaging the nitrogenous bases.
The chemical bonds that the enzymes cleave can be reformed by other enzymes known as ligases, so that restriction fragments carved from different chromosomes or genes can be spliced together, provided their ends are complementary.
Many of the procedures of molecular biology and genetic engineering rely on restriction enzymes. The term restriction comes from the fact that these enzymes were discovered in E. coli strains that appeared to be restricting the infection by certain bacteriophages.
Restriction enzymes therefore are believed to be a mechanism evolved by bacteria to resist viral attack and to help in the removal of viral sequences. They are part of what is called the restriction modification system. The 1978 Nobel Prize in Medicine was awarded to Daniel Nathans, Werner Arber and Hamilton Smith for the discovery of restriction endonucleases, leading to the development of recombinant DNA technology.
The first practical use of their work was the manipulation of E. coli bacteria to produce human insulin for diabetics. The ability to produce recombinant DNA molecules has not only revolutionized the study of genetics, but has laid the foundation for much of the biotechnology industry. The availability of human insulin (for diabetics), human factor VIII (for males with hemophilia A), and other proteins used in human therapy all were made possible by recombinant DNA.
Types of Restriction Enzymes:
Restriction enzymes are classified biochemically into three types. These are designated as Type I, Type II, and Type III. A major type of Type II enzymes are sometimes referred to as Type IV enzymes.
Type I and III systems, both the methylase and restriction activities are carried out by a single large enzyme complex. Although these enzymes recognize specific DNA sequences, the sites of actual cleavage are at variable distances from these recognition sites, and can be hundreds of bases away. Both require ATP for their proper function.
Type I restriction enzymes produce DNA cleavage following translocation of the DNA, which makes them important molecular motors. The cleavage of DNA appears to occur after blockage of the translocation activity (often following collision with another translocating Type R-M enzyme, but also due to other factors).
These enzymes read the methylation status of their recognition sequence, compare the methylation status of two adenines within the recognition sequence, and if both adenines are un-methylated (a signal that the DNA is non-host DNA), the enzyme undergoes a conformational switch that turns the enzyme into a molecular motor and endonuclease.
However, if either one of the adenines is methylated (a signal that the DNA is host DNA) then the enzyme acts as a maintenance methylase and methylates the other adenine. In Type II systems, the restriction enzyme is independent of its methylase, and cleavage occurs at very specific sites that are within or close to the recognition sequence. The vast majority of known restriction enzymes are of type II, and it is these that find the most use as laboratory tools. They produce discrete bands during gel electrophoresis, and are useful for DNA analysis and gene cloning. The first to be discovered and utilized was EcoRI, which is staggered and its recognition sequence is 5′-GAATTC-3′.
Type II enzymes are further classified according to their recognition site. Most type II enzymes cut palindromic DNA sequences, while type IIa enzymes recognise non-palindromic sequences and cleave outside of the recognition site, and type lIb enzymes cut sequences twice at both sites outside the recognition sequence. Type lIs enzymes cleave the DNA at a considerable offset from the recognition sequence. The most common type II enzymes are those like HhaI, HindIII and NotI that cleave DNA within their recognition sequences.
Restriction enzymes usually occur in combination with one or two modification enzymes (DNA-methyltransferases) that protect the cell’s own DNA from cleavage by the restriction enzyme.
Modification enzymes recognize the same DNA sequence as the restriction enzyme that they accompany, but instead of cleaving the sequence, they methylate one of the bases in each of the DNA strands. The methyl groups protrude into the major groove of DNA at the binding site and prevent the restriction enzyme from acting upon it.
Together, a restriction enzyme and its “cognate” modification enzyme(s) form a restriction-modification (R-M) system. In some R-M systems the restriction enzyme and the modification enzyme(s) are separate proteins that act independently of each other. In other systems, the two activities occur as separate subunits, or as separate domains, of a larger, combined, restriction-and-modification enzyme.
Restriction Enzymes as Tools:
Recognition sequences typically are only four to twelve nucleotides long. Because there are only so many ways to arrange the four nucleotides—A,C,G and T–into a four or eight or twelve nucleotide sequence, recognition sequences tend to “crop up” by chance in any long sequence. Furthermore, restriction enzymes specific to hundreds of distinct sequences have been identified and synthesized for sale to laboratories.
As a result, potential “restriction sites” appear in almost any gene owr chromosome. Meanwhile, the sequences of some artificial plasmids include a “linker” that contains dozens of restriction enzyme recognition sequences within a very short segment of DNA. So no matter the context in which a gene naturally appears, there is probably a pair of restriction enzymes that can cut it out, and which will produce ends that enable the gene to be spliced into a “plasmid”.
Another use of restriction enzymes can be to find specific SNPs. If a restriction enzyme can be found such that it cuts only one possible allele of a section of DNA (that is, the alternate nucleotide of the SNP causes the restriction site to no longer exist within the section of DNA), this restriction enzyme can be used to genotype the sample without completely sequencing it. The sample is first run in a restriction digest to cut the DNA, and then gel electrophoresis is performed on this digest.
If the sample is homozygous for the common allele, the result will be two bands of DNA, because the cut will have occurred at the restriction site. If the sample is homozygous for the rarer allele, the sample will show only one band, because it will not have been cut. If the sample is heterozygous at that SNP, there will be three bands of DNA. This is an example of restriction mapping.