Let us make an in-depth study of the DNA replication:- Learn about: 1. Basic Features of DNA Replication 2. Mechanism of DNA Replication 3. Meselson and Stahl Experiment 4. Enzymes of DNA Replication 5. Formation of Replication Forks & Replication Bubbles and Others.
Central Dogma:
Genetic material is always nucleic acid and it is always DNA except some viruses. DNA is the storehouse of genetic information. This information is in the form of nucleotide sequence called genetic code. This information is copied and transcribed into RNA molecules. This information (genetic code) is for specific sequence of amino acids. The RNA then synthesizes proteins, which are specific sequence of amino acids, by a process called translation. In 1956 Francis Crick called this pathway of flow of genetic information as the Central Dogma.
Both transcription and translation are unidirectional. Proteins never serve as template for RNA synthesis. But sometimes RNA acts as a template for DNA synthesis (reverse transcription), Example is RNA viruses (HIV virus).
Contents
DNA Replication:
Genetic information present in double stranded DNA molecule is transmitted from one cell to another cell at the time of mitosis and from parent to progency by faithful replication of parental DNA molecules. DNA molecule is coiled and twisted and has enormous size. This imposes several restrictions on DNA replication. DNA molecule must be uncoiled and the two strands must be separated for the replication process.
Basic Features of DNA Replication:
All genetically relevant information of any DNA molecule is present in its sequence of bases on two strands. Therefore the main role of replication is to duplicate the base sequence of parent DNA molecule. The two strands have complementary base pairing. Adenine of one strand pairs with thymine of the opposite strand and guanine pairs with cytosine. This specific complementary base pairing provides the mechartism for the replication.
The two strands uncoil and permanently separate from each other. Each strand functions as a template for the new complementary daughter strand. The base sequence of parent or old strand directs the base sequence of new or daughter strand. If there is adenine in the parent or old strand, complementary thymine will be added to the new strand. Similarly, if there is cytosine in the parent strand, complementary guanine will be copied into the new daughter strand. Maintenance of integrity of genetic information is the main feature of replication.
Mechanism of DNA Replication:
Mechanism of DNA replication is the direct result of DNA double helical structure proposed by Watson and Crick. It is a complex multistep process involving many enzymes.
1. Initiation:
It involves the origin of replication. Before the DNA synthesis begins, both the parental strands must unwind and separate permanently into single stranded state. The synthesis of new daughter strands is initiated at the replication fork. In fact, there are many start sites.
2. Elongation:
The next step involves the addition of new complementary strands. The choice of nucleotides to be added in the new strand is dictated by the sequence of bases on the template strand. New nucleotides are added one by one to the end of growing strand by an enzyme called DNA polymerase. There are four nucleotides, deoxyribrnucleotide triphosphates dGTP, dCTP, dATP, dTTP present in the cytoplasm.
3. Termination:
All the end termination reactions occur. Duplicated DNA molecules are separated from one another.
The purpose of DNA replication is to create two daughter DNA molecules which are identical to the parent molecule.
DNA Replication is Semi-Conservative:
Watson and Crick model suggested that DNA replication is semi-conservative. It implies that half of the DNA is conserved. Only one new strand is synthesized, the other strand is the original DNA strand (template) that is retained. Each parental DNA strand serves as a template for one new complementary strand.
The new strand is hydrogen bonded to its parental template strand and forms double helix. Each of these strands of the double helix contains one original parental strand and one newly formed strand.
Meselson and Stahl Experiment:
Mathew Meselson and Franklin Stahl proved experimentally that parental strands of a helix are distributed equally between the two daughter molecules. They made use of the heavy isotope 15N as a tag to differentially label the parental strands. E. coli was grown in a medium containing 15N labeled NH4Cl.
In this way both strands of DNA molecules were labeled with radioactive heavy isotope 15N in their purines and pyrimidines. Therefore both strands were heavy or HH DNA. The bacteria were then transferred into a medium containing the common non-radioactive nitrogen 14N, which is a light medium. It was found that after one cell division daughter molecules had one 15N strand the other 14N strand. So this is a hybrid molecule, a heavy light of HL.
After the second cell division, out of four molecules, two DNA molecules contained UN(LL). The other two were hybrid molecules (HL). This proves that during replication, one parent strand is conserved and the other new strand is synthesized. Thus DNA replication is a semi-conservative process.
Enzymes of DNA Replication:
The enzymes which take part in replication are able to copy DNA molecules which may contain millions of bases. They perform this function with utmost accuracy and at high speed, even though DNA molecule is highly compact and is bound with proteins. Maintenance of integrity of genetic information is the main feature of replication.
In E. coli, two main enzymes that take part in polymerization are DNA polymerase I and DNA polymerase III. Of these DNA polymerase III is the main enzyme involved in replication. Polymerization involves addition of new nucleotides to a growing strand.
In addition, there is an enzyme DNA polymerase II which takes part in DNA repair. DNA polymerase IV and DNA polymerase V have also been discovered. DNA polymerases are capable of adding 1000 nucleotides per second. The speed of DNA synthesis is known as processivity.
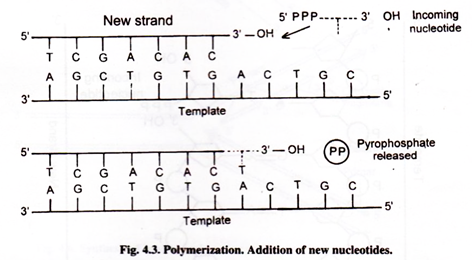
Polymerization:
Nucleotide monophosphates, which are building blocks of DNA, cannot be added to the growing strand of DNA. The DNA polymerase can act only on deoxyribose triphosphate nucleotides. They are dATP, d GTP, d CTP and dTTP. Triphosphate nucleotides possess high-energy phosphate bonds.
DNA polymerase I and DNA polymerase III can add new nucleotides at the 3′-OH end of the growing strand. Inorganic pyrophosphate is released. Hydrolysis of pyrophosphate is the driving force for DNA synthesis. Phosphodiester bond is formed between 3′-end of growing strand and 5′-end of first phosphate of incoming nucleotide.
Template:
The two DNA strands separate and each acts as a template for the formation of new strand. Polymerization-reaction is dictated by a template strand according to the base pairing rules where if adenine is present in the template, thymine is added to the new strand and guanine pairs with cytosine.
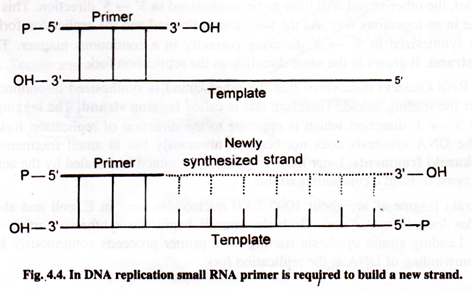
Primer:
DNA polymerase needs primer to synthesize new strand on it. Primer is a small strand segment which is complementary to the template. It has a free 3′-OH end to which a new nucleotide can be added. In this way part of the new strand is already in place. Primer is hydrogen bonded to the template to form primer: template junction. Primer is short nucleotide strand (oligonucleotide). The primer for both the new strands is RNA primer. The RNA primer is synthesized by an enzyme, primase.
DNA polymerase moves along the template adding nucleotides. Thus DNA is synthesized by extending the 3′-end of the primer. Afterwards, primer is removed by DNA polymerase I and RNAse H.
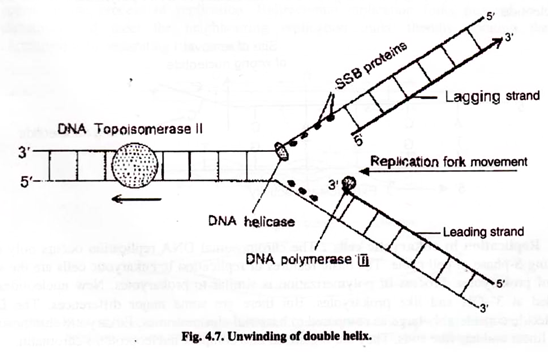
Leading strand requires only a single RNA primer. On the other hand, discontinuous synthesis of lagging strand requires primer for each Okazaki fragments. For synthesis of lagging strand hundreds of Okazaki fragments with their associated RNA primers are required.
In this way two conditions are necessary for DNA synthesis. They are a template and a primer with 3′-OH end.
DNA Synthesis takes Place in 5′ → 3′ Direction Only:
A new strand of DNA is always synthesized in 5′ → 3′ direction. The free 3′-end enables it to be elongated. Because the two strands are antiparallel, orientation of new growing strand is opposite to the template strand.
DNA Replication is Discontinuous in One Strand:
If the synthesis is to proceed in 5′ → 3′ direction, only one strand can be synthesized in correct 5′ → 3′ direction. As the strands are antiparallel, the other strand will have to be synthesized in 3′ 5′ direction.
This constraint is overcome in an ingenious way. As the two strands unwind and the replication fork grows, one strand is synthesized in 5′ 3′ direction correctly in a continuous manner. This is called leading strand. It grows in the same direction as the replication fork.
Mr. Reiji Okazeki discovered that the other strand is synthesized discontinuously and a little after the leading strand. Therefore this is called lagging strand. The lagging strand also grows in 5′ 3′ direction which is opposite to the direction of replication fork. In lagging strand, the DNA synthesis does not occur continuously but in small fragments, which are called Okazaki fragments. Later these fragments are joined and sealed by the action of DNA ligase enzyme to form a continuous strand.
Okazaki fragments are about 1000-2000 nucleotides long in E. coli and about 100-200 nucleotides long in eukaryotes. Both the strands begin the synthesis starting on a primer segment. Leading strand synthesis starting on a primer proceeds continuously keeping pace with the unwinding of DNA at the replication fork.
Each Okazaki fragment is synthesized on a short RNA primer. DNA polymerase III binds to RNA primer and adds deoxyribonucelotides. Lagging strand proceeds in opposite direction from the fork movement.
The primer ribonucleotides are removed and replaced by deoxyribornucleotides and then joined. The removal of RNA primer is done by exonuclease activity of DNA polymerase I.
Unwinding of Double Helix:
The first step of DNA replication is the unwinding parent double helix molecule so that each strand acts as a template for the new strand. Unwinding mechanism is very complex. Hydrogen bonds between two strands are broken. This is achieved by enzymes called DNA helicases which move along DNA and separate the strands.
DNA helicases bind to the lagging strand template. Strand separation create topological stress like the one produced if the two ends of a coiled rope are pulled apart fc- separation. This forms supercoils in the unreplicated double helix in front of the replication fork. This stress is removed and strands are separated by the action of DNA topoisomerase Topoisomerase I introduces cut or nick in one DNA strand.
Topoisonmerase II remove supercoils by causing double stranded breaks. Some parts of DNA molecule have circle linked as a chain. This structure is called catenane. DNA gyrase is capable of decatenating two circles, thus separating them. In this way these enzymes open up and unwind the DNA helix.
In order to prevent the formation of hydrogen bonds again between two separated strand- the two single strands are coated by Single Strand Binding Proteins (SSB proteins), which stabilize the separated strands.
Semi-discontinuous Replication:
Leading strand is synthesized continuously but lagging strand is synthesized discontinuously. This is called semi-discontinuous replication.
Replication is Highly Accurate:
Replication takes place with an extraordinary accuracy In spite of all this, errors do occur and wrong nucleotides are added. Approximately one mistake occurs in every 101” nucleotides added. But mechanisms do exist for the repair of DNA molecule. In fact DNA is the only molecule for which repair mechanisms exist.
Proof Reading or Editing Functions:
DNA polymerase I is a very versatile enzyme. In addition to its ability to polymerize, it also performs repair function. Sometimes a wrong nucleotide is added at the 3′-OH end which fails to base pair with the complementary base on the template strand. Then the polymerization process is stopped. The enzyme moves backward and removes the wrong base by degrading DNA.
Then the polymerization activity resumes and chain growth starts again by adding correct base. It removes only the most recent error. This process is known as 3′ → 5′ exonuclease activity and the enzyme is called proof reading exonuclease. This editing function gives a second chance to DNA polymerase to add correct nucleotide.
Replication in Eukaryotic Cells:
The chromosomal DNA replication occurs only once during S-phase of cell cycle. The basic features of replication in eukaryotic cells are the same as of prokaryotes. Process of polymerization is similar to prokaryotes. New nucleotides are added at 3′-OH end like prokaryotes. But there are some major differences.
The DNA molecule considerably large as compared to bacterial chromosomes. Eukaryotic chromosomes are linear and has free ends. They are organized into complex nucleoproteins chromatin.
Replication in eukaryotes is considerably faster. Replication is completed in three minutes in embryonic cels of drosophilla. Chromosomes of higher organisms have multiple origins of replication and all replication forks proceed bidirectionally.
Eukaryotic DNA has repeated units of replication called replicons. Enormous number of replication units require large number of polymerase enzyme molecules. An animal cell has 20000 – 60000 molecules of polymerase ∝.
Eukaryotes have several types of polymerase enzymes. The main three enzymes are DNA polymerase ∝ DNA polymerase δ and DNA polymerase є. Replication is initiated by DNA polymerase ∝ and DNA polymerase δ and є bring about rapid polymerization because of their high processivity.
Eukaryotic replication also synthesizes end structures or telomeres.
Formation of Replication Forks and Replication Bubbles:
Initiation of replication occurs within the double helix and rarely at the end. Opening up of DNA molecule creates replication bubble. Replication bubble progresses in the form of replication fork in one direction in the case of unidirectional replication and in both directions in bi-directional replications.
Bidirectional Replication:
In circular DNA of bacteria and linear DNA of eukaryotes, DNA replication proceeds bidirectionarlly starting from a fixed origin of replication. The two replication forks move in opposite directions. Bidirectional replication may have multiple replication forks. They speed up the process of replication.
Eukaryotic chromosomes are very long. In order to speed up the process of replication, a chromosome may have thousands of points of origin of replication (O). They tremendously speed up the process of replication. Bidirectional replication forks proceed in opposite directions and meet the neighbouring replication units, thereby opening the entire chromosome by separating two strands.
Unwinding and Replication of Circular Double Helix DNA of E. Coli:
Most of the bacteria have double stranded circular DNA with no free ends. This poses a problem of unwinding at the time of replication. Replication originates at one point and the two replication forks proceed in opposite directions. The advancing replication forks meet at a point opposite to the point of origin thus opening up the coiled DNA molecule. This is called θ (theta) model of replication.
But the unwinding is a very complex mechanism as the two strands are coiled. The two advancing replication forks make the remaining entire un-replicated portion of DNA overwound. Thus, the un-replicated portion becomes so tightly coiled that the advancing replication forks are not able to advance further.
This is because of positive supercoiling of the un-replicated portion. In E. coli an enzyme called DNA gyrase produces negative super coiling, thus removing the positive supercoiling. This leads to unwinding of the entire double helix circular chromosome of the bacteria.
Newly Synthesized Eukaryotic DNA Immediately Forms Nucleosomes:
Before the replication takes place, the DNA disentangles itself from the nucleosomes. After replication, newly synthesized DNA immediately rejoins the octomers of histone proteins to form nucleosomes.
Large amounts of histones are synthesized during .S’-phase of interphase. Old histones are not lost. Old histones are present on both daughter chromosomes. During replication, nucleosomes are broken down into their components and later reassembled into nucleosomes.
Summary:
Central Dogma:
DNA is the storehouse of genetic information. This information in the form of nucleotide sequence called genetic code. This information is transcribed onto RNA which translate this information into sequence of amino acids (Protein). Francis Crick called this central dogma
DNA Replication:
Genetic information present in double stranded DNA molecule is transmitted from one cell to another cell and to progeny by faithful replication of DNA molecules. The main role of replication is to duplicate the base sequence of parent DNA molecule. The two strands uncoil and permanently separate from each other. Each strand
functions as a template for the new complementary daughter strand.
The base sequence of parent or old strand directs the complementary base sequence of new or daughter strand. Replication includes steps initiation, elongation and termination. DNA replication is semi- conservative. Out of two strands formed, one old or parental strand is retained and the other view strand is synthesized. This was experimentally proved by Meselson and Stahl in E. coli. Main enzyme involved in replication is DNA polymerse III. Other enzymes involved are DNA polymerase I, II, IV and V.
Polymerization:
Four kinds of nucleotides are building blocks of DNA. The DNA polymerase acts on dATP, dGTP, dCTP, dTTP. These new nucleotides are added one-by-one at 3′-OH end of the growing strand. DNA polymerase needs a primer to synthesize new strand. Small RNA primer hydrogen bonds with the template. This primer provides free 3′- OH end to add new nucleotides. DNA synthesis takes place in 5′ → 3′ direction.
Therefore only one strand which grows in the direction of replication fork is synthesized in 5′ → 3′ direction. This is called leading strand and is synthesized continuously. The other strand is synthesized in small fragments in 5′ → 3′ direction. These fragments are called Okazaki fragments. Later these fragments are joined and sealed by DNA ligase enzyme. This strand is called lagging strand and is synthesized discontinuously.
Unwinding of DNA Double Helix:
As each strand acts as a template for new strand, the two strands must unwind. Hydrogen bonds between two strands are broken by enzymes called DNA helicases. DNA topoisomearases also help in unwinding. Single strand binding proteins (SSB proteins) bind the unwound single strands to prevent the formation of hydrogen bonds again.
Editing Function:
Sometimes wrong bases are added at the 3′-OH end. DNA polymerase I enzyme performs the repair function. It moves backward 3′ → 5′ direction and removes the last base. This is called exonuclease function. Then DNA synthesis resumes is 5′ → 3′ direction.
Replication in Eukaryotic Cells:
The basic features of replication are same in prokaryotes and eukaryotes except some difference. Polymerization is similar in both cases. Eukaryotic chromosomes are much longer and have free ends. Polymerase enzymes are DNA polymeraze α DNA polymerase δ and DNA polymerase є.
Formation of Replication Forks and Replication Bubbles:
Opening up DNA molecule creates replication bubbles which progress in the form of replication fork. Replication fork progresses in one direction in case of unidirectional replication and in both directions in bi-directional replication. Eukaryotic chromosomes are very long, so there may be thousands of points of origin of replication. This speeds up the process of replication tremendously.
Newly synthesized eukaryotic DNA immediately forms nucleosomes.