Expression Vectors: 5 Things to Know About:- The five things are:
(1) RNA Expression (2) Protein Expression(3) Screening cDNA Expression Libraries with Antibodies (4) Fusion Protein Expression and (5) 1 -β-Galactosidase and Blue or White Selection.
Frequently it is the product of the cloned gene, rather than the gene itself, that is of primary interest— particularly when the protein has commercial, therapeutic, or research value.
With an increased understanding of the fundamentals of DNA, RNA, and protein metabolism and their regulation, investigators can now manipulate cells to express cloned genes in order to study their protein products.
Expression vectors are engineered so that any cloned insert can be transcribed into RNA, and, in many instances, even translated into protein. cDNA expression libraries can be constructed in specially designed vectors derived from either plasmids or bacteriophage l.
Proteins encoded by the various cDNA clones within such expression libraries can be synthesized in the host cells, and if suitable assays are available to identify a particular protein, its corresponding cDNA clone can be identified and isolated. Expression vectors designed for RNA expression or protein expression, or both, are available.
Thing # I. RNA Expression:
A vector for in vitro expression of DNA inserts as RNA transcripts can be constructed by putting a highly efficient promoter adjacent to a versatile cloning site. Figure 4.14 depicts such an expression vector. Linearized recombinant vector DNA is transcribed in vitro using SP6 RNA polymerase. Large amounts of RNA product can be obtained in this manner; if radioactive ribonucleotides are used as substrates, labelled RNA molecules useful as probes are made.
Thing # II. Protein Expression:
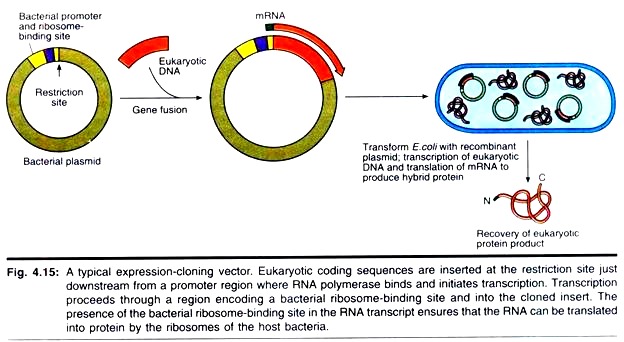
Because cDNAs are DNA copies of mRNAs, cDNAs are uninterrupted copies of the exons of expressed genes. Because cDNAs lack introns, it is feasible to express these cDNA versions of eukaryotic genes in prokaryotic hosts that cannot process the complex primary transcripts of eukaryotic genes. To express a eukaryotic protein in E. coli, the eukaryotic cDNA must be cloned in an expression vector that contains regulatory signals for both transcription and translation.
Accordingly, a promoter where RNA polymerase initiates transcription as well as a ribosome binding site to facilitate translation are engineered into the vector just upstream from the restriction site for inserting foreign DNA. The AUG initiation codon that specifies the first amino acid in the protein (the translation start site) is contributed by the insert (Fig. 4.15).
Strong promoters have been constructed that drive the synthesis of foreign proteins to levels equal to 30% or more of total E. coli cellular protein. An example is the hybrid promoter, ptac, which was created by fusing part of the promoter for the E. coli genes encoding the enzymes of lactose metabolism (the lac promoter) with part of the promoter for the genes encoding the enzymes of tryptophan biosynthesis (the trp promoter) (Fig. 4.16).
In cells carrying ptac expression vectors, the ptac promoter is not induced to drive transcription of the foreign insert until the cells are exposed to inducers that lead to its activation. Analogs of lactose (a β-galactoside) such as isopropyl- β-thiogalactoside, or IPTG, are excellent inducers of ptac. Thus, expression of the foreign protein is easily controlled. The bacterial production of valuable eukaryotic proteins represents one of the most important uses of recombinant DNA technology.
For example, human insulin for the clinical treatment of diabetes is now produced in bacteria. Analogous systems for expression of foreign genes in eukaryotic cells include vectors carrying promoter elements derived from mammalian viruses, such as simian virus 40 (SV40), the Epstein-Barr virus, and the human cytomegalovirus (CMV).
A system for high-level expression of foreign genes uses insect cells infected with the baculovirus expression vector. Baculoviruses infect lepidopteran insects (butterflies and moths). In engineered baculovirus vectors, the foreign gene is cloned downstream of the promoter for polyhedrin, a major viral-encoded structural protein, and the recombinant vector is incorporated can lead to accumulation of the foreign gene product to levels as high as 500 mg/l.
Thing # III. Screening cDNA Expression Libraries with Antibodies:
Antibodies that specifically cross-react with a particular protein of interest are often available. If so, these antibodies can be used to screen a cDNA expression library to identify and isolate cDNA clones encoding the protein. The cDNA library is introduced into host bacteria, which are plated out and grown overnight, as in the colony hybridization scheme previously described.
DNA binding nylon membranes are placed on the plates to obtain a replica of the bacterial colonies. The nylon membrane is then incubated under conditions that induce protein synthesis from the cloned cDNA inserts, and the cells are treated to release the synthesized protein.
The synthesized protein binds tightly to the nylon membrane, which can then be incubated with the specific antibody. Binding of the antibody to its target protein product reveals the position of any cDNA clones expressing the protein, and these clones can be recovered from the original plate. Like other libraries, expression libraries can be screened with oligonucleotide probes, too.
Thing # IV. Fusion Protein Expression:
Some expression vectors carry cDNA inserts cloned directly into the coding sequence of a vector- borne protein-coding gene (Fig. 4.17). Translation of the recombinant sequence leads to synthesis of a hybrid protein or fusion protein. The N-terminal region of the fused protein represents amino acid sequences encoded in the vector, whereas the remainder of the protein is encoded by the foreign insert.
Keep in mind that the triplet codon sequence within the cloned insert must be in phase with codons contributed by the vector sequences to make the right protein. The N-terminal protein sequence contributed by the vector can be chosen to suit purposes. Furthermore, adding an N-terminal signal sequence that targets the hybrid protein for secretion from the cell simplifies recovery of the fusion protein.
A variety of gene fusion systems have been developed to facilitate isolation of a specific protein encoded by a cloned insert. The isolation procedures are based on affinity chromatography purification of the fusion protein through exploitation of the unique lig- and-binding properties of the vector-encoded protein (Table 4.2).
Thing # V. 1 -β-Galactosidase and Blue or White Selection:
One version of these fusion protein expression vectors places the cloning site at the end of the coding region of the protein β-galactosidase, so that among other things the fusion protein is attached to β-galactosidase and can be recovered by purifying the β-galactosidase activity.
Alternatively, placing the cloning site within the β-galactosidase coding region means that cloned inserts disrupt the β-galactosidase amino acid sequence, inactivating its enzymatic activity. This property has been exploited in developing a visual screening protocol that distinguishes those clones in the library that bear inserts from those that lack them.
Cells that have been transformed with a plasmid-based β-galactosidase expression cDNA library (or infected with a similar library constructed in a bacteriophage λ-based β-galactosidase fusion vector) are plated on media containing 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside, or X-gal (Fig. 4.18).
X-gal is a chromogenic substrate, a colourless substance that upon enzymatic reaction yields a coloured product. Following induction with IPTG, bacterial colonies (or plaques) harbouring vectors in which the β-galactosidase gene is intact (those vectors lacking inserts) express an active β-galactosidase that cleaves X-gal, liberating 5-bromo-4-chloro- indoxyl, which dimerizes to form an indigo blue product.
These blue colonies (or plaques) represent clones that lack inserts. The P-galactosidase gene is inactivated in clones with inserts, so those colonies (or plaques) that remain “white” (actually, colourless) are recombinant clones.