The following points highlight the two main causes of ploids. The causes are: 1. Aneuploidy 2. Euploidy.
Ploids: Cause # 1. Aneuploidy:
The individuals carrying chrornosome numbers other than somatic or diploid (2n), which are not true multiples of basic chromosome number or genome are called heteroploids or aneuploids. Thus aneuploidy results due to an addition of loss of less than a complete set of chromosomes to or from a diploid set.
Thus there are two classes of aneuploids:
(i) Hyperploids which result due to addition of one or more chromosomes to a diploid set, and
(ii) Hypoploids which develop due to loss of one or more chromosomes from a diploid set.
Causes or Origin of Aneuploids:
1. Both the classes of aneuploids arise from an abnormal distribution of chromosomes during anaphase of meiosis I. Because of irregular chromosome distribution at the poles, one daughter cell receives one or more extra chromosomes and the counterpart lacks in one or more chromosomes.
This type of abnormal situation arises when one or more pairs of chromosom (bivalents) fail to separate or disjoin and move to one spindle pole as such, this is called non-disjunction.
The gametes having extra chromosome, when fuse with normal haploid gametes result in hyperploids and those containing one or more chromosomes less than a haploid number if unite with normal haploid gametes produce hypoploids.
2. Loss of individual chromosome in meiosis or mitosis leads to the formation of nuclei with hypoploid chromosome number.
3. Irregularities in the segregation of chromosomes during meiosis in polyploids.
4. Multipolar mitosis with irregular chromosome distribution to the daughter cells.
Heteroploids or aneuploids are arranged in several groups. They are listed in Table 23.1 and Fig. 23.1.
1. Monosomic (2n-1):
An organism lacking one chromosome of a diploid complement is called monosomic (2n -1). Suppose in an diploid set is formed of 4 pairs of chromosomes. In this, by chance one chromosome is missing. So in this case one may find 3 normal pairs of chromosomes and one singlet chromosome.
This should, therefore, be called as monosomic. Since monosomies, as well as nullisomic and trisomy affect only one chromosome of the haploid complement, there may be expected as many different type of monosomes as are the chromosomes in the haploid set.
It some plants, monosomies are not viable. In Nicotiana tabaccum with basic chromosome number 24, Clausen and his co-workers have found all 24 possible monosomies as expected and have noted they were much different from one another.
In wheat (Triticum vulgare) haploid number ‘n’ is 21. So one may expect that 21 monosomics of different types should be available. Sears (1948) obtained all the 21 possible monosomics in bread wheat (Triticum vulgare), but they did not differ much from the normal diploids.
The monosomies are very rare. However, viable monosomies have been studied in Drosophila, tobacco, Datura, maize, tomatoes and many other organisms.
Monosomies show abnormal type of meiosis. They have chromosome constriction 2n-1, so they produce two kinds of gametes; some with n and others with n – 1 chromosomes. The odd chromosome, which has no homologue to pair tends to pass at random to one pole during meiosis I or may divide, as in mitosis, into two chromatids which move to opposite poles.
Frequently, however, it acts as a laggard at anaphase and is not included in any of the daughter nuclei and subsequently lost. For this reason the gametes having chromosomes occur less frequently than the gametes of (n – 1) type (e.g., wheat produces about 75% n- 1 gametes).
In plants, the nuclei with a missing chromosome seldom survive. The n – 1 gametophytic generation fails, presumably because the complete absence of certain genes that occur on the missing chromosome means the failure of certain biochemical reactions which are accomplished in presence of the enzymes ‘formed by these genes.
Thus, monosomic analysis makes it clear what genes are found on the particular chromosome or linkage group.
2. Trisomic (2n + 1):
An organism containing two complete genomes plus one extra chromosome is termed trisomic (2n+1) In the normal process of meiosis I, sometimes chromosomal pairs separate in such a way that one member of each pair goes to one daughter nucleus and other member goes to other daughter nucleus.
But very rarely one pair of chromosomes fails to disjoin and finally it moves as such to one spindle pole. So half of daughter cells receive an extra chromosome and half of them lose one.
In this way gametes with (n -1) and (n + 1) chromosomes will develop. If by chance the gamete with (n + 1) chromosomes fuses with normal gamete carrying ‘n’ chromosomes, the resulting zygote will have n+1 or 2n + I chromosomes and it will produce trisomic organism.
Gametes with (n – 1) chromosomes on fusion with gamete carrying ‘n’ chromosomes will give rise to monosomic (2n -1) zygote. Many of the mutations in Oenothera noted by De Vries are trisomies. Trisomies are widespread in nature and have been extensively studied in Datura, maize, tomatoes, wheat, tobacco and Drosophila.
Blakeslee and Belling (1924) announced a mutant type in Datura having 25 chromosomes – rather than normal 24 chromosomes (2n = 24). In that, at first meiotic metaphase one of the 12 pairs was found to have an extra member, i.e., there were 11 normal pairs and one trisome.
The extra chromosome in the trisomic is either completely homologous or partially homologous to a certain chromosomes of the standard complement and, accordingly, the trisomies may be classified into Primary, Secondary and Tertiary trisomies.
Primary Trisomies:
When the extra chromosome is completely homologous to one pair of homologues in the diploid chromosome compliment and, therefore, one chromosome is represented three time in an otherwise diploid complement, it is primary trisomy.
Such homology permits formation of a trivalent or one bivalent and rod shaped univalent during prophase of meiosis I. The trivalent may be of different configurations as shown in Fig. 23.3.
The number of different possible trisomies for a given karyotype corresponds to the number of chromosomes in each chromosome set. Since in Datura, there are 12 chromosome pairs with different genie composition, and the triplication of every chromosome pair is possible, 12 distinguishable trisomies can be expected in it.
Blakeslee and his associates have succeeded in producing all 12 different trisomies as expected (Fig. 23.2).
These trisomies were grown in Blakeslee’s garden and all were found to have distinct phenotypic features which could easily be attributed to extra dose of one chromosome to each of the 12 individual sets of chromosomes.
Trisomies show irregular distribution of chromosomes during anaphase I of meiosis. From a trisomic cell the two kinds of gametes; some with ‘n’ chromosomes and some with (n +1) chromosome, would be expected after meiosis.
Recessive genes present on the extra chromosomes in trisomic complexes will express themselves less frequently than in normal diploid plants because comparatively more wild type alleles are present to check their expression.
It has been observed by geneticists in Datura that extra chromosome enters the offspring through female gamete because the egg tolerates of one chromosome without any loss of viability. The pollen with extra dose of chromosome is not viable. Only normal pollen with haploid chromosome number succeeds in fertilizing the egg.
Primary trisomic individuals exhibit several specific features which are quite different from those seen in the wild types. The chromosomal association in primary trisomic is show in fig 23.4. The additional traits associated with the extra chromosome give the plants extra dose of all the genes that are already contained in the normal duplicated chromosomes.
The viability and fertility are usually lower than those of diploids from which they arise.
(ii) Secondary Trisomics:
When the trisomic contains an extra chromosome that has two identical arms (isochromosome), it is said to be secondary trisomic. For each chromosome two types of secondary chromosomes are possible.
At first meiosis either a ring trivalent which is rare in primary trisomic (diagnostic configuration) or a bivalent and one U-shaped ring resulting from the internal pairing of the univalent with identical arms may be formed (Fig. 23.4).
Secondary trisomies may occur among the progeny of normal diploid individuals but their best source is among the progeny of individuals with one or more univalent chromosomes in which misdivision of centromere takes place.
Tertiary Trisomies:
In these, the extra chromosome is the result of reciprocal translocation between two standard chromosomes. The ends of extra chromosome are homologous with the ends of two different chromosomes. The tertiary trisomies are identified by the presence of a chain of three les. The tertiary trisomic individual has two chromosome segments represented three times each an otherwise diploid complement.
The tertiary trisomies occur regularly and possibly exclusively in the progenies of translocation heterozygotes by 3: 1 segregation of four chromosomes which are partially homologous in such heterozygotes and may form a quadrivalent during prophase I of meiosis.
Segregation of genes included in a primary trisomic association differs from that of genes found in any of the other chromosomes of the complement. For a particular locus (A/a), a trisomic may have the genotype AAA (triplex), AAa (duplex), Aaa (simplex) or aaa (nulliplex).
If one of the three chromosomes is not eliminated, selfing or intercrossing of trisome would theoretically result in 25% tetrasomics, 50% trisomies and 25% disomies (diploid) progeny. But this expectation is rarely realized because of irregular meiosis and non-functioning of n+ 1 gametes.
A regular chromosome distribution at meiosis I (two to one pole and one to the other) in genotypes AAa and Aaa would lead to expected gametic ratio 1 AA : 2Aa : 2A : la (dominant: recessive = 5:1) and 2Aa: 1 aa : 1A: 2a (dominant: recessive = 1:1) respectively. If the male gametes possessing the additional chromosome (n + 1) are non-viable as in many plants and chromatid segregation is absent, the following genotype proportions are expected in the progenies of selfed or intercrossed trisomies of genotypes, AAa and Aaa (Table 23.2):
The observed genotypic ratio in the progenies may deviate considerably from the above theoretical expectation. If instead of chromosome segregation, chromatid segregation takes place and the crossing over takes place between centromere and locus, then the expected gametic ratio for the genotype AAa will be 6AA: 8Aa: laa :l 0A:5a instead of lAA: 2Aa: 2A :1a.
3. Double Trisomies (2n + 1 + 1):
Sometimes an organism receives two complete genomes plus two extra chromosomes. This is called double trisomic. The extra chromosomes are homologues of two different chromosomal pairs in normal diploid set. Thus two different chromosome pairs in the normal diploid set will have one additional homologue each with them and thus they form two trisomes.
The phenotypic expression of genes on extra chromosomes in trisomies will be deeper than in normal diploids.
4. Tetrasomic (2n + 2):
Aneuploid nuclei which have normal diploid genome and one extra pair of chromosomes are called tetrasomics (2n + 2). In this one pair of chromosomes in the diploid set is reduplicated due to additional chromosome pair. Phenotypic expression of the genes in tetrasomics may be deeper than the effects found for the genes in corresponding trisomies.
In meiosis, the four homologues of tetrasomic set often tend to form a quadrivalent (Fig. 23.5). If disjunction by twos is regular, a fair genetic system will operate. Generally quadrivalents are not formed and disjunction (separation) is not always regular. Nevertheless, tetrasomics are more regular than trisomics in meiosis.
5. Nullisomic (2n-1):
In these both the homologues of a particular chromosome set are somehow lost so that a chromosomal pair is completely missing from the normal 2n set. The phenotypic expression of such a plant, when compared with that of normal plant, will indicate the number of genes contained in the missing chromosomes and the respective characters they govern.
Nullisomics are generally non-viable like the monosoes. In maize and wheat, several nullisomics have been reported. Nullisomics are generally obtained by selfing monosomies. When the gametes containing n-1 chromosomes fuse, they form nullisomics.
Ploids: Cause # 2. Euploidy:
The euploidy (Gr. eu., even or true; ploid, unit or set) refers to a state in which a cell or an individual contains to one or more complete chromosome set or sets. The individuals having an integer number of chromosome sets or genomes are called euploids. Euploids are classified into haploids, diploid and polyploids as detailed in Table 23.3:
Haploids or Monoploids:
Monoploids or haploid organisms contain single genome in their cells, i.e., they contain one member of each kind of chromosome. The haploid chromosome number (n) is usually found in gametes of diploid plants and animals and this is unusual for a somatic cell of higher plant.
Monoploidy has been reported in several cases, both plants and animals. Viruses and bacteria whose genome consists of single linkage structure are haploids.
Majority of lower plants, particularly thallophytes and bryophytes exist in monoploid condition. In higher plants, haploids develop as a result of parthenogenesis of haploids eggs. In Datura stramonium, Oryza sativa, Oenothera, Triticum, Zea mays and many other plants, haploidy has been noted. In some animals also, haploids are common. The males of bees, wasps and other hymenoptera are normally haploids.
Characters of Haploids:
1. These plants are usually weaker and smaller than the diploids, but in pepper the haploids are as healthy as normal diploid plants.
2. Leaves are generally small.
3. Plants are of low viability.
Cytology of Haploids:
High degree of sterility has been observed in these plants. It is so only because meiosis is irregular. As the chromosomes in haploid set do not have their homologues, the pairing of chromosomes is not possible and the chromosomes are found as univalent’s at metaphase I of meiosis. So neither the eggs nor the pollens are formed.
However, if meiotic process succeeds at all, the univalent chromosomes are found scattered all over the cell. They may constitute restitution nucleus including all chromosomes and may thus give rise to gametes having a complete haploid set of chromosomes.
In haploid Datura stramonium dyads are produced by microsporocytes which directly enter second meiosis and such haploid spores are functional. Sometimes the distribution of chromosomes to the two poles is irregular and the gametes so formed are highly non-viable (i.e., they die soon).
In haploid animals, reduction division does not occur during spermatogenesis. Normal gamete is possible if by chance all the chromosomes pass to one pole during the irregular meiosis.
According to Kimber and Riley (1963) the haploids may be classified as follows on the basis of ploidy status of their parents:
Monoploids may arise in several ways. In plants, they arise by parthenogenetic development of embryo from unfertilized eggs.
Monoploids and haploids occur spontaneously in maize in low frequencies. They may be induced from pollen grains through callus or embryoid production and by chromosome elimination through interspecific hybridization as in Hordeum jubatum x H. vulgare.
Production of haploids can be induced by using pollen incapable of effecting fertilization such as pollen from a related but incompatible species or the pollen inactivated by exposures to high dosages of x-rays or the pollen made in-viable by prolonged storage prior to pollination.
Application of Monoploids in Breeding:
Monoploids are generally sterile and are of no use in agriculture directly. They do not undergo segregation and carry a single set of genes. So they are genetically pure. Thus, they can be used experimentally to a good advantage in crop improvement.
(j) They are used for developing homozygous diploid lines. In maize, extensive inbreeding is necessary to select lines for hybrid seed production. Monoploids provide a short cut way for getting homozygous diploids which are then tested for their value in the production of hybrid seeds.
The normal diploids can be obtained by treating monoploid tissue with colchicine solution.
(ii) They may be useful in isolation of mutants because the mutant allele, even if recessive, expresses itself in M, generation due to a single dose of gene in somatic tissues. Haploid mutants can be used for developing homozygous diploid mutant line in a single generation.
(iii) Pure autotetraploid varieties of certain economic crops like potato can be produced by doubling the chromosome number of haploids to tetraploid level.
(iv) It is easier to get desirable haploid derived diploid lines than to get a desirable zygote derived lines.
Diploids:
Most common type of euploid is the diploid which has two complete genomes. As mentioned earlier this arrangement is nearly universal among animals and common in plants. These are normal in their behaviour, so they are not discussed here in detail.
Polyploids:
Polyploidy is commonly met within plant world. In animals, it is indeed rare. About one-half of total species of flowering plants are polyploids. Among certain families the proportion of to diploids is higher than 50 per cent.
About two-third species of all grasses are polyploids. The case of polyploidy may be found in many other common plants e.g., Chrysanthemum, Solanum, Brassica, Wheat. In the genus Chrysanthemum, the basic number of chromosome is 9.
In this genus species are known with 36,54,63,72 and 90 chromosomes. In wheat (Triticum sp.) the basic chromosomes is 9. In this, several varieties are known with 14, 28 and 42 chromosomes which are diploid tetraploid and hexaploid respectively. In nicotiana with basic chromosome number 12, the varieties with 24, 48 and 72 chromosomes are known. These are all polyploids.
Similarly the genus solanam, in which basic chromosome number is 12, the varieties with 24, 48 and 72 chromosomes. In Rose, the basic number is 5. In this, diploid, triploid, tetraploid, hexaploid and octaploids have 10, 15, 20, 30’and 40 chromosomes respectively (Fig. 23.6).
In animals, the polyploids are rare on account of more sex balance than in plants. In animals, addition of les generally gives rise to intersexes which are: sterile (i.e. do not reproduce).
Sterility is virtually always with departure from diploid number in animals, animals which can reproduce partheno-genetically (e.g. Anemia salina) develop polyploid races. It is so because parthenogenesis enables these animals to escape from the of anomalous gametes.
Two Types of Polyploids:
Two factors could account for the origin of polyploids from diploids:
I. Doubling of the chromosome number in reproductive tissue capable of giving rise to gametes.
2. Failure of reduction in the chromosome number during the formation of gametes. On the basis of their origin, the polyploids are of the following two types.
1. Autopolyploids:
The individuals containing more than two genomes or sets of homologous chromosomes per cell are called autopolyploids. More scientifically, in these organisms, multiple genomes are identical or nearly so. Autopolyploidy arises due to the failure of anaphase resulting in the duplication of genomes.
For example, if a diploid species has two identical sets of chromosomes or genomes (AA), the autotriploid will have three similar genomes (AAA) and the autotetraploid will have four such sets of homologous chromosomes (AAAA) per cell (Fig. 23.7).
2. Allopolyploids:
The individuals in which the multiple genomes are not alike are called allopolyploid. In allopolyploids, different genomes are derived from two or more distinct species by hybridization and contain the structurally and genetically different sets of chromosomes of the particular parents, each set being present more than once.
Suppose, there are two different species, one with genomes AA and the other with BB. A cross between these two species will produce a hybrid AB. If the genomes A and B of the hybrid are multiplied spontaneously or artificially, so that the individual contains two or more sets of each genome, it is called allopolyploid (Fig. 23.7).
If the duplication of different genomes or sets of chromosomes takes place only once, it is referred to as amphidiploidy or allotetraploidy and the resulting polyploid is called allotetraploid or amphidiploid (e.g., AA, BB). If the multiplication is taking place more than once (i.e., higher multiplication) it is called allopolyploidy. It results in allopolyploids like allohexaploids, allooctaploids, etc.
Autopolyploidy:
Autopolyploids are not very common in occurrence and are of little evolutionary consequence. Polyploidy appears very seldom in animals but in plants many species are known to be represented by polyploids.
Morphological Features of Autopolyploids:
The morphological features of polyploids may vary in different species. Some general features are listed here as follows:
(i) In many cases autopolyploidy lead to increase in general vigour and vegetative growth and so they are often larger than their diploid counterparts. In some cases, however, autopolyploids are smaller and weaker than diploids.
(ii) Polyploids have large cell sizes than diploids. The guard cells are larger and the frequency of stomata per unit area is lower in the polyploids than in diploids.
(iii) Pollen grains in polyploids are larger than in corresponding diploids.
(iv) The cells of polyploids contain more chloroplasts than those of diploids.
(v) Polyploids are slower in growth and have thicker and larger leaves as compared to diploids.
(vi) The flowering occurs later in polyploids than in diploids.
(vii) Allopolyploids generally show reduced fertility due to high irregularities during meiosis which causes genotypic imbalance leading to physiological disturbances.
(viii) The flowers in polyploids are showy and larger than those in diploids.
(ix) The flowers and fruits per plant in autopolyploids are usually less in number than in diploids.
(x) Autopolyploids have, in general lower dry mass than diploids, e.g., in turnip (Brassica rapa) and cabbage (B. oleracea). This shows that increased size of polyploid may not represent increased dry matter.
(xi) Different species have different levels of optimum ploidy as for example, 3n in sugarbect (Beta vulgaris), 8 – 10n in Phleum pratense grass. Autopolyploidy is much successful in species with low chromosome numbers and in cross-pollinated species.
Crops grown vegetatively are more likely to succeed as’ polyploids than those grown by seeds. The autopolyploids which are derived from diploid species differ only quantitatively in phenotype from their diploid parents.
The quantitative phenotypic difference, if present, can be due to the following causes:
(a) The increased size of nuclei and cells can differentially influence the gene activity (Goldschmidt, 1937).
(b) The increase in number of alleles per locus associated with autopolyploidy can modify the expressivity of the alleles.
(c) If the diploid form is heterozygous for the concerned allelic pairs, an increase in the number of chromosome sets may lead to modifications of the dominance relationships of the alleles.
Cytological Behaviour of Autopolyploids:
The cytological behaviour of chromosomes vary with the level of ploidy. In auto-triploids, a variable number of trivalents, bivalents and univalents may be observed in the meiotic cells during prophase I. At anaphase I, separation of chromosomes of trivalents and distribution of univalents are irregular.
All possible gamete types with chromosome numbers between haploid to diploid may result which may produce a range of aneuploid progeny, e.g., trisomies, monosomies, tetrasomics, nullisomics, double trisomies and so on. Autotriploids are highly sterile.
In autotetraploids, there are four sets of homologous chromosomes. During the prophase of meiosis I, chromosomes of each set are present as quadrivalents or otherwise. 2 bivalents or one univalent and one univalent or four univalents.
The pairing of chromosomes derived from one genome is called autosyndesis. Autotetraploids have somewhat higher fertility because the chances of normal pairing are increased, but the fertility is in all cases lower than that in diploids.
In most part, the autopolyploids with even number of haploid sets are more fertile than those with an odd number of sets. In the progeny of autotetraploids, the majority of plants are tetraploids, but some plants showing variable numbers of chromosomes may also be produced.
There is a considerable evidence that the fertility of autotetraploids can be improved through selection. This has been achieved in some cases such as maize, rye, rice, bajra etc.
Segregation of Genes in Autopolyploids:
The meiotic segregation of genes in autopolyploids is more complex than in diploids. The number of alleles of each gene is represented according to the ploidy level of the individual and gametes containing more than one allele of each gene (homo or heterozygotic) may be produced.
According to the number of dominant and recessive genes at a particular locus, the genotype of an autotetraploid may be quadriplex and monoallelic (AAAA or A4), triplex and biallelic (AAAa or A3a), duplex and biallelic (AAaa or A2 a2), simplex and biallelic (Aaaa or Aa3) and nulliplex and monoallelic (aaaa or a4). Autopolyploids show the so called polysomic inheritance.
The segregation of genes in autopolyploids is affected by factors which play no essential role in diploid. Among such factors are the number and position of chiasmata in the multivalents, the distance between particular locus and centromere, the behaviour of homologues in multivalent associations during anaphase I and the presence of univalents.
In autotriploids, the actual segregation ratios correspond, in principle, with those of trisomies but, as a rule, these ratios are strongly affected by irregularities of chromosome distribution and by chromosome elimination. Therefore, it is difficult to predict segregation pattern of genes in autotriploids.
In autotetraploids, if it is assumed that the four homologous chromosomes are distributed to poles in 2: 2 during anaphase I, theoretical segregation ratios for various autotetraploid genotypes of a locus may be calculated. Theoretically calculated segregation ratios which do not take into consideration, crossing over between the locus concerned and the centromere are referred to as chromosome segregation.
The segregation ratios which take into account regular crossing over between the concerned locus and centromere and thereby modified segregation of heterozygous loci are referred to as “chromatid segregation” (double reduction). When the locus of gene is very close to centromere, crossing over would not occur between the centromere and gene locus.
In such a case the two sister chromatids of each chromosome are attached to the same centromere and would move to same pole during anaphase I of meiosis. The relative frequencies of the gamete types and zygote types of different autotetraploid genotypes after selfing and reciprocal crossing, the identical genotypes for pure “chromosome segregation” are presented in the following Table 23.4.
Table 23.4 Frequencies of the gamete types and zygote types of autotetraploid genotypes for pure Chromosome segregation.
If it is assumed that the two sister chromatid segments with identical alleles of one chromosome are distributed after crossing over, the corresponding distribution of chromatids into the same meiotic product will modify expectations of gamete and zygote classes (Table 23.5).
The extent of chromatid segregation actually observed in autotetraploid depends upon the locus of gene and crossing over frequency between the concerned locus and centromere, frequency to multivalent formation, types of orientation and distribution of chromosomes in multivalent associations.
The segregation ratios of different genotypes of autotetraploids generally lie between those theoretically expected for pure chromosome segregation and pure chromatid segregation.
Origin of Autopolyploids:
The autopolyploidy develops directly or indirectly through chromosome doubling. It may occur spontaneously or may be induced by spindle positions. It is the result of the formation of a restitution nucleus in somatic cell (somatic polyploidization) or in reproductive tissues (generative or gametic polyploidization).
Some Specific Examples of Autopolyploids:
Triploids:
Such plants have 3 genomes per nucleus and they are commonly designated as 3n plants. Autotriploids occur in a number of plants, e.g., Oenothera, Datura, rose, rice and many others. They are usually formed as a result of fertilization between a diploid (2n) and a haploid gamete (n), (2n gamete + n gamete = 3n zygote) (Fig. 23.8).
Diploid gametes are produced by normal tetraploids in meiosis or in sectors of otherwise diploid organisms where automatic doubling of somatic chromosome number (i.e., of 2m) has taken place. Somatic doubling may be spontaneous or may be induced.
Characters:
(i) Autotriploids are more vigorous than normal diploids, they are more leafy and show tendency towards perenniality.
(ii) Sometimes floral abnormalities may be observed.
(iii) Pollengrains, stomatal guard cells and wood cells of xylem in triploids are larger than those of diploids.
(iv) Plants are highly sterile and seeds are formed rarely. In nature, seed propagated triploid plants are ordinarily rare because of irregularity at meiosis.
Generally the autotriploids occurring in nature are propagated by vegetative means. This is advantageous in horticulture, specially for ornamental plants, e.g., Chrysanthemum, roses, Dahlias, etc., and also in the production of seedless fruits. Some of the commercially important fruit plants, as for example, apples, pears, bananas, grapes, oranges, guavas, pineapples are triploid.
Meiosis:
In autotriploid it is irregular. The centromeres of three homologous chromosomes have no way to orient themselves so as to give equivalents at the two poles. In this condition, the only possibility is that the components of trivalent separate in such a way that two chromosomes go to one side and third goes to the other.
If it is so then the meiotic products may contain either ‘n’ or ‘2n’ chromosomes or any number in between.
This is not true irrespective of whether three homologous chromosomes align either as trivalent or as one bivalent and one univalent. Therefore, the gametes arising from triploids have unbalanced genomes. In fact, probability of formation of haploid and diploid gametes is very rare.
Autotetraploids (4n):
Autotetraploids have four similar genomes per nucleus.
They may originate in one of the following ways:
(i) By fusion of two diploid gametes, and
(ii) They may result from the duplication of somatic chromosomes (2 n) followed by failure of mitotic division.
The resulting nucleus contains four copies of each chromosomes, instead of the usual two. If this tetraploid perpetuates itself through normal mitosis, the increased chromosome number may become established in a group of cells or tissues within the body of organism. Plants capable of vegetative propagation may be manipulated to produce pure tetraploids.
Characters:
(i) Autotetraploids usually show marked phenotypic variations, great adaptability and sometimes marked disease resistance. They are commercially more valuable than corresponding diploids,
(ii) Some are larger and more deeply coloured than diploids and sometimes they bear large seeds and fruits,
(iii) They have larger cells with correspondingly larger nuclei than do the diploids,
(iv) The stomatal guard cells and epidermal cells are larger and arranged in different patterns in tetraploids than those in diploids,
(v) Flowers are larger and more showy in tetraploids than those in diploids,
(vi) High content of vitamin C (ascorbic acid) have been reported in tetraploids, and
(vii) The tetraploids show tendency towards perenniality and may show low fertility.
Meiosis:
It may be normal or abnormal. When four homologous align in tetravalents, normal diploid gametes are formed. Sometimes, two bivalent or one trivalent and one univalent may also occur. In these cases, products of meiosis are irregularly formed. Incomplete genomes in gametes may result in total sterility.
Allopolyploidy:
Allopolyploids have genomes from two or more genetically distinct species, each genome being present more than once in a nucleus. The occurrence of allopolyploids has been reported in many plant species including some crops, as for example in Raphanobrassica, Triticale, Rosa, Primula kewensis, Spartina townsendii, Papaver, Iris versicolor, Grepis, etc.
Origin and Production of Allopolyploids:
The allopolyploids existing in nature were produced spontaneously by chromosome doubling in the natural hybrids of two distinct species belonging to the same genus or different genera (Fig. 23.9).
The chromosome doubling might have occurred in the somatic cells (Somatic allopolyploidization) of the hybrid due to irregular mitosis resulting in the formation of allopolyploid sector either in the apical meristem or in the auxiliary buds.
Such sectors of allopolyploid tissue in the buds would have produced allopolyploid branches. Sexual progeny from such allopolyploid branches possibly resulted in stable natural allopolyploids.
The other alternative way by which allopolyploids could arise might be the formation and fusion of unreduced functional gametes (generative allopolyploidization).
In the artificial production of allopolyploids from distant hybrids both the processes, somatic and generative polyploidization have been followed and many important allopolyploids have been developed.
A very important case of allopolyploidy is Raphanobrassica which was experimentally evolved by the Russian geneticist G.D. Karpechenko who made intergeneric cross between radish (Raphanus sativus) and cabbage (Brassica oleracea) (Fig. 23.10). Although these plants were distantly related, they were enough alike to be crossed successfully with each other. Both had 9 pairs of chromosomes.
The diploid hybrid Raphanobrassica had 18 chromosomes; 9 chromosomes from radish and 9 chromosomes from Brassica parent. But it could not perpetuate itself largely because of failure of pairing between unlike chromosomes during meiosis.
When chromosomes of sterile Raphanobrassica were doubled by artificial means, a fertile polyploid Raphanobrassica was produced with 36 chromosomes; 18 of radish and 18 of cabbage.
Experimental production of allopolyploids is achieved by doubling the chromosome number of distant hybrids with the help of colchicine or some other agents. Thus, the homologous chromosomes could pair themselves and the regular meiosis resulted normal gametes, each with 18 chromosomes.
This pairing of chromosome is called autosynapsis, as actually chromosomes of two genomes are pairing with their respective replicas. This experiment had theoretical significance since it provided a method by which fertile interspecific hybrids could be produced. This also suggests the possibility of incorporating the desirable genotypes of two different species into a new polyploid species.
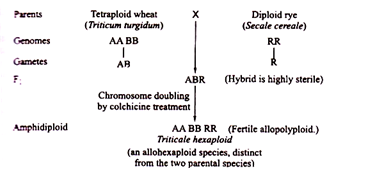
Triticale is another artificially produced allopolyploid produced by crossing tetraploid or hexaploid wheat with rye (Secale cereal)
Triticale derived from tetraploid wheat parents have been the most successful species as compared to those derived from hexaploid wheats. Presently triticales are being grown commercially in some countries, e.g., Canada, Sweden, Mexico, etc. The triticales have been found to be as good in production as any high yielding variety of wheat.
Morphological Features of Allopolyploids:
Since the allopolyploids contain genomes of two or more genetically distinct species, they generally combine the morphological and physiological characteristics of parent species. Generally, it is difficult to predict the precise combination of characters that are likely to develop in the allopolyploid species.
For example, Raphanobrassica was synthesized with the aim of combining the root character of radish (Raphanus sativus) and the leaf character of cabbage (Brassica oleracea) but Raphanobrassica has been found to behave in quite opposite direction as it shows leaves like those of radish and root like that of cabbage.
But, the reverse is also true in some of the cases, as for example, Triticale has combined the desirable features of parent species, i.e., the hardiness of Secale cereale (rye) and yielding potency of wheat (Triticum species).
In general, allopolyploids may be more vigorous than diploids but there are many exceptions to it.
Since the allopolyploids show different natural distribution than their progenitor diploid species, it appears that they have different adaptability than the parent species. Rarely the allopolyploids may displace the parental species from their natural habitats. Allopolyploid plants, as a whole, possess a good deal of robustness. Fruits are characteristically somewhat larger than those of diploid parents.
Another remarkable feature of allopolyploids is the occurrence of apomixis (replacement of sexual reproduction by various types of asexual reproduction). Many species of grasses are sexually sterile allopolyploids which reproduce by apomictic methods.
Cytological Behaviour of Allopolyploids:
In allopolyploids or amphiploids each chromosome of the original allodiploid or amphihaploid (e.g., AB) is represented twice. The original amphihaploid are sterile hybrids, chromosomes of which behave, more or less, like those of haploids. By formation of allopolyploid, fertile derivatives are obtained.
Meiotic behaviour of allopolyploids is governed by the homology relationships of chromosome sets they possess.
There are two types of allopolyploids:
(i) Some allopolyploids form only bivalents and no multivalents or univalents are observed during meiosis. In such a case every chromosome finds one identical partner with which it pairs. Such an allopolyploid breeds true for its genotype and there is no segregation.
(a) The presence of homologous chromosome sets in more than double dose (e.g., AAAA BB), and
(b) The introduction of partially homologous (= homoeologous) chromosome sets.
The segregation of allopolyploids is consequence of behaviour of chromosomes in multivalent associations in both cases. Generally, extreme genetical imbalance occurs among segregants of allopolyploids with numerous complex multivalents in meiosis resulting in reduced fertility.
On the basis of meiotic behaviour of chromosomes of different sets, Stebbins (1945) classified allopolyploids as follows:
1. Genome allopolyploids:
In these the, chromosomes pairing is exclusively in the form of bivalents during meiosis. Pairing is restricted to exactly homologous chromosomes (homogenetic pairing) which may be consequence of the striking structural dissimilarities between the parental chromosome sets or may result from the activity of specific genes which prevent the pairing of partially homologous chromosomes.
The amphihaploid or allodiploid parent of such allopolyploid is usually completely sterile. The fertility is fully restored with the production of the genome allopolyploid derivative. Once established, this derivative is usually completely isolated by sterility barriers from its nearest relatives and behaves like a new species.
In genome allopolyploid, only identical chromosomes of the similar genomic sets pair, behaving as they do in diploid organisms. This type of chromosome pairing is called autosyndatic pairing or autosyndasis. For this reason allotetraploid with complete autosyndatic pairing is called amphidiploid (=double diploid)
2. Segmental allopolyploids:
In these, the chromosomes form bivalent and multivalent associations. The parental chromosome sets combined in the diploid hybrid in this case are partially homologous and correspond in a considerable number of segments permitting “heterogenetic pairing”. The pairing of partially homologous chromosomes from different genomes is referred to as allosyndesis.
The term describes pairing of any kind between A chromosome and B chromosome which are homologous in AA BB tetraploid. In allosyndatic pair the chromosomes have one or more small segments that are homologous. Thus, the sterility of diploid hybrid parent of such an allopolyploid is not very pronounced.
In contrast to genome allopolyploid, the segmental alloploids show more or less extensive segregation; the segregants being highly dis-balanced genetically and sterile. Hence such polyploids are genetically unstable. Through segregation, they may produce stable segregants, which in regard to their chromosome, constitution may be autoploid, genome allopolyploid or stable segmental allopolyploid.
3. Autoallopolyploids:
In these allopolyploids, chromosomes may form bivalents and multivalents like those in segmental polyploids, because homologous chromosomes are present in more than double dose. Polyploids of this type are possible from hexaploid level upward and combine characteristics of auto and allopolyploidy.
The fertility of allopolyploids can be improved through hybridization and selection. Selection improves chromosome pairing behaviour and through recombination’s new gene combinations develop that improve the physiological and cytological behaviour of allopolyploids.