Lecture notes on chromosome. After reading these notes you will learn about: 1. Introduction to Chromosome 2. Meaning of Chromosome 3. Number and Size 4. Structure and Morphology 5. Molecular Organization 6. Mechanism of Chromosome Condensation 7. Special Types 8. Karyotype Concept 9. Heterochromatin.
Contents:
- Notes on Introduction to Chromosome
- Notes on the Meaning of Chromosome
- Notes on the Number and Size of Chromosome
- Notes on the Structure and Morphology of Chromosome
- Notes on the Molecular Organization of Chromosome
- Notes on the Mechanism of Chromosome Condensation
- Notes on the Special Types of Chromosomes
- Notes on the Karyotype Concept of Chromosome
- Notes on Heterochromatin in Chromosome
Note # 1. Introduction to Chromosome:
The knowledge regarding the chemistry of the chromosome goes as far as 1874 when Miescher, on chemical analysis of nuclei of various animals, reported the presence of a protein component and another substance – nuclein in Salmon sperm, now known as nucleic acid.
Later, the works of different authors led to the understanding of chemical structure of chromosomes having the continuous frame work of protein and nucleic acid, latter being responsible for hereditary stability.
Deoxyribonucleic acid, the principal component of chromosome of nucleus, in combination with protein, forms the nucleoprotein component of the chromosomes. The initial concept of a chromosome being composed of a skeleton of basic protein surrounded by deoxyribonucleic acid, has undergone significant changes.
It is now known, that both basic and acidic protein along with deoxyribonucleic acid enter into the chromosome constitution. The ribonucleic acid too, according to some authors, appears at certain stages of the cycle, principally in the synaptonemal complex.
The presence of both types of protein as well as DNA has been confirmed through enzyme digestion studies utilizing deoxyribonucleic for DNA, and trypsin and pepsin for digesting basic and non-basic protein respectively.
However, from the studies on animal organs, the term residual’ chromosome was adopted which is supposed to be the fundamental skeleton of the chromosome. This fundamental skeleton which, undergoes spiralization during divisional cycle, reveals the presence of acidic proteins and RNA in addition to DNA.
It is supposed that the nucleohistone fraction forms the matrix over the residual chromosomes. The former can be removed by sodium chloride treatment and fractionation, keeping the fundamental structure intact. However, such residual structure, claimed to be the fundamental skeleton, has not been located in all organisms.
Note # 2. Meaning of Chromosome:
The chromosomes are the bearers of hereditary material and vehicles of transmission from cell to cell and generation to generation. The genes which are the unit basis of heredity are located inside the chromosomes. Each chromosome is thus a giant complex molecule made up of smaller but less complex molecules the genes which are arranged in linear sequence in the chromosome thread, inside the nucleus.
The presence of chromosome was first demonstrated by Strasburger (1875) and described as chromosome (chroma-colour; soma-body) by Waldeyer (1888) due to their marked affinity for basic dyes. The term ‘chromosome’ was first coined by Baranetsky as the principal component in the nucleus and is responsible for maintenance of hereditary stability.
In the lower organism, in the prokaryote with no distinct nuclear membrane, the chromosome is represented in the DNA alone. This structure is referred to as genophore, (sensustricto), as compared to the complex organized structure – the chromosome, in higher forms of life or eukaryotes.
The structure of genophore being at the level of molecular dimension is beyond the resolution of light microscope, and can be analysed at the ultra-structural level under electron microscope. The chromosomes of higher organism on the other hand, can be observed under light microscope and are made up of nucleic acid and proteins – both basic and non-basic.
Chromosome research has been greatly enriched through physical, chemical and molecular studies, coupled with basic cytological and genetic analysis. Such a synthetic approach has given a deeper understanding of structure and behaviour of chromosomes and modality of the genetic control on every aspect of cell metabolism.
Note # 3. Number and Size of Chromosome:
The number and size of chromosome remain constant for a particular species. Each and every cell of an organism contains the same number and types of chromosomes which enables each gene to exert the influence. The constancy in the number throughout the body, is maintained by a type of cell division otherwise termed as mitosis which involves equational separation of chromosomes.
Each daughter cell contains chromosome complements of both the parents and is designated as diploid. The halving of chromosome number is achieved through another type of cell division, otherwise termed as meiosis or reduction division which leads to the formation of cells with half the chromosome number in resulting gametes both male and female and are designated as haploid.
The original diploid number is restored following sexual reproduction, which involves the union of both male and female gametes, leading to fertilization. The reduction division or halving of chromosome number leading from diploidy to haploidy, is accompanied by pairing and interchange of segments (crossing over) between paternal and maternal chromosomes (homologous chromosome pair).
Though the number of chromosomes remains constant for a particular species, it varies from species to species. The lowest and highest chromosome number recorded in plants are Haplopappus gracilis (2n=4) of Composite and Ophioglossum (2n ≥ 1200) of Pteridophyta respectively (Fig. 3.1).
The chromosome size as studied from mitotic metaphase may be as short as 0.25 µm in fungi to as long as 30 µm in Trillium; the breadth ranging from 0.2µm to 2 µm.
Note # 4. Structure and Morphology of Chromosome:
Chromosome is the principal component of the nucleus of higher plants. It contains genes in linear order, responsible for maintaining hereditary stability. In the cell, the chromosomes appear as coiled threads in the early part of nuclear division, which become condensed at later stage.
Chromonema and Chromomere:
The basic structure of chromosome is a thread-like structure termed chromonema by earlier authors. The chromonema thread, on the other hand, is beaded in appearance, the beads being termed as chromomeres. The understanding of such beaded thread-like structure of chromosomes as visualized by earlier authors, has undergone much refinements in later years.
The present concept of chromosome structure is regarded as fibrillar, made up of fibrils, folded several times to give a diameter of 10 pm thickness (Fig. 3.2A). The structure appears as coiled at the beginning of cell division which undergoes spiralization and condensation up to a stage, followed by successive de-spiralization and de-condensation (Fig. 3.2B).
It is a continuous DNA-protein fibre in which condensed and de-condensed segments alternate. The single condensed segment of a chromosome of higher organism is comparable to the entire DNA thread of a microbe.
The chromonema and chromonemic component of the chromosome can, however, be resolved under the light microscope at certain stages of division, such as prophase, where the structure is not fully spiralized. The chromonema represents coiled thread and is the permanent component of chromosome.
The chromomere represents a higher order of compaction and contains submicroscopic beads, the nucleosome which can be resolved only at the ultra-structural level.
Chromatid:
At the light microscopic level, observation of chromosome is dependent on the use of basic dyes. The chromosome structure is analysed principally from the metaphase, i.e., the mid-phase of cell division where the chromosomes are highly condensed and spiralized and as such responds strongly to basic dyes.
Each chromosome at metaphase is made up of two symmetrical chromatids which are held together by centromere and become separated at anaphase.
Constriction-Primary and Secondary:
By the usual methods of fixation and staining, the chromosomes at the dividing stages appear as somewhat cylindrical bodies constricted at one region or more along their lengths (Fig. 3.3). The position of these constrictions are constant characteristics of any particular chromosome of the complement.
They are frequently referred to as primary and secondary constrictions, the former representing a region of the chromosome involved in movement during division, while the latter occurs only in certain chromosomes being associated with the production of nucleoli (nucleolar zone).
The primary constriction, referred to as centromere is obviously an essential part, since chromosomes which lack the centromere cannot move to the poles during mitosis. The secondary constriction, referred to as NOR, responsible for the organization of nucleolus.
Centromere and Kinetochore:
The centromere is a specialized region of the chromosome associated with its movement along the spindle to the two poles during anaphase. It may be localized on a particular part of the chromosome the primary constriction region or it may be diffuse in which the entire chromosome expresses the property of spindle attachment.
The centromere forms:
(i) Final point of adhesion for the sister chromatids during metaphase to anaphase transition,
(ii) Attachment point for mitotic spindle fibres, as well as,
(iii) Site for motor protein to mediate chromosome movement.
The localized centromere appears basically as small de-stained gap during metaphase. This appearance is due to the presence of two blocks of heterochromatin at the two sides of the centromeric apparatus, which exhibit allocycly. They are de-stained at metaphase and stained at interphase to form brightly stained bodies or pro-chromosomes in the interphase nucleus.
The position of the centromere has been taken as the criterion for designating chromosomes in different categories. Based on the relative position of the centromere, chromosomes are described as metacentric, sub-metacentric, acrocentric and telocentric (Fig. 3.4A). If the centromere is towards the middle, dividing the chromosome in two equal arms, the chromosome is known as metacentric.
Similarly, sub-metacentric and acrocentric are terms used to designate chromosome having constrictions at the sub-median position or towards the end respectively. If the centromere is located at the end of the arm, it is known as telocentric. On the basis of position of centromere, the chromosomes look ‘V shaped, ‘L’ shaped, ‘J’ shaped, ‘V shaped respectively during anaphasic movement (Fig. 3.4B).
During a normal division, centromeric chromomeres and threads divide lengthwise so that an identical replica is transmitted to each daughter chromosome. However, under certain conditions, transverse division has been observed, referred to as misdivision or bursting of the centromere by Darlington (1939) in species of Nicandra. The resultant daughter chromosomes show terminal centromeres.
On the basis of the number of centromere, chromosome may be mono-centric (one centromere – usual), dicentric (two centromeres – as in wheat), polycentric (many centromeres – as in Ascaris) or acentric (without centromere – does not survive).
The diffuse centromeres do not show any single attachment point and show parallel movement of two chromatids throughout the length on the spindle. The diffuse nature of the centromeres of Luzula has also been confirmed through irradiation of chromosomes, where the broken fragments behave as chromosomes with centromeres. As such, in this genus, increase in chromosome number arises through fragmentation.
Kinetochore is the microtubule implantation site to which spindle microtubules are attached (Fig. 3.5A). Electron microscopic studies have given further details of the structure of the kinetochore. These are trilaminar cup or plate like structures of 0.20-0.25 nm diameter on either side of the centromeric constriction.
In cross section it has 3 layers:
(i) An outer layer of dense material of 10 nm thick
(ii) A middle layer of lower density,
(iii) An inner dense layer in contact with the underlying chromatin fibres.
Chemically kinetochore is made up of specialized DNA-protein complexes. The DNA sequences representing the centromere contain: information specifying the assembly of kinetochore. In certain higher plants, such as species of Lilium and Tradescantia, long blocks of repetitive sequences have been located around the centromeres of several species.
In general, the centromeric DNA-protein complex is resistant to nuclease digestion in vitro. Centromeres are shown to suppress meiotic recombination in some systems. 
Many plants have clusters of highly repetitive sequences at or near their centromere. In a few plant species, tandem repeats are being specifically located at or around the centromere. Different types of proteins too have been localized in the centromere.
The centromeric DNA sequences, isolated from different organisms, have not been found to be essentially conserved. The centromere of budding yeast contains three DNA sequence elements (CDEI, CDEII, CDEIII) of 250 bp long to which centromeric binding proteins (CBF- centromeric binding factor) are attached (Fig. 3.5B).
In fission yeast, the centromeric region contains at least four classes of repetitive DNA elements. Viz., L, K and B sequences flanking a core sequence. The sequences essential to centromeric function in yeast are about 130 bp long and rich in AT pairs; the satellite centromeric DNA is found to consists of thousands of tandem copies of one or few short sequences of 5-10 bp long.
The centromeric region in higher plants and animals is much more complex and includes both unique and repetitive DNA sequences. All primates contain the satellite centromeric DNA of 171 bp long tandem repeats with a small functional sequence of 17 bp long, called CENP-B box.
The centromeres of plants like rice, maize, wheat contain two different conserved repeats which often show homology to CENP-B box of mammals or CDEII of yeast. Different types of proteins have been detected to be localized in the centromere and their role in centromere structure is also distinct.
NOR and Satellite:
In addition to the centromere, otherwise known as primary constriction, which has a role in chromosome movement, some of the chromosomes possess another constriction, termed secondary constriction (Fig. 3.6).
This constriction, if is located almost at the end of the chromosome arm so that the terminal segment assumes a dot-like appearance, the term satellite is used (Fig. 3.7) and the chromosome is called SAT chromosome. The word satellite, though often widely used, is in a strict sense is the end segment of the chromosome arm.
The satellite thread, i.e., continuous thread of the chromosome representing the connecting link between the so-called satellite and the main chromosome arm is often visible. The secondary constriction or the satellite segment is responsible for the organization of the nucleolus, hence called Nucleolus Organizing Region or NOR.
As this segment contains genes coding for 18 S and 28 S ribosomal RNA, it is necessary for ribonucleoprotein metabolism. In addition, the chromosome in certain cases, may have more than one secondary constriction – known as supernumerary constrictions.
Telomere:
Telomeres are nucleoprotein structures and occur at the ends of the chromosomes. They are enlarged terminal ends of chromo-sortie and are heterochromatic as observed by a different staining cycle. They perform several vital functions including end protection.
Telomeres enable the chromosomes to distinguish between normal chromosome ends and breaks, so that the cycle can be delayed to repair’ chromosome breaks.
The protection of the chromosome ends is mediated through a special mechanism involving telomerase a reverse transcriptase. This compensates for the inability of DNA polymerase to replicate chromosome completely. Normal DNA replication mechanism cannot lead to complete telomere replication as they yield a blunt DNA end and the other end having 3′ overhang.
The stability and complete replication of the telomeres of higher organisms indicate that they have certain common features. Telomeric DNA, comprising the extreme molecular ends of chromosomes, consists of simple tandemly repeated sequences, characterized by clusters of G residues in one strand. An overall asymmetric strand composition results in G-rich and complementary C-rich strands.
The 3′ end of each strand of the duplex chromosomal DNA molecule is the G-rich telomeric strand and it forms a 3′ terminal overhang, 12 to 16 nucleotides in length, protruding from the duplex (Fig. 3.8A). Each species has a characteristic telomeric repeat sequence. Limited sequence variations are found in some species. However, widely divergent species can have the same telomeric repeat unit as well.
Polymerization of TTGGGG is achieved by adding into the telomeric oligonucleotide primer at the 3′ end of a G-rich strand, independent of an exogenously added nucleic acid template. Each telomerase synthesizes its species-specific G-rich strand sequence (Fig. 3.8B).
The model of telomere serves as a molecular clock controlling the life-span. The telomeric DNA in eukaryotic cell is progressively lost in successive generations on account of both incomplete replication and strand-specific exonuclease activity. In case the loci is not made up, the telomere continues to shorten with generation doubling. Thus in absence of telomeric activity, chromosome shortens at each division.
Note # 5. Molecular Organization of Chromosome:
The chromosomes of higher organism have been shown to be made up of fibrils, approximately 20-30 Å in diameter, folded several times to yield a diameter of 10 pm thickness. It represents a continuous DNA-protein fibre. Regarding the number of fibrils along the length of chromosome, multi-stranded concept was proposed by different scientists (Steffensen, Ris) earlier.
Later it has been claimed with experimental evidences that the single molecule runs along the entire length of the chromosome from telomere to telomere uninterrupted through the centromeric region. Thus the chromosome structure has been demonstrated to represent a uninemic skeleton (single stranded concept).
Several models have further explained the manner of association and folding of DNA with protein in chromosome.
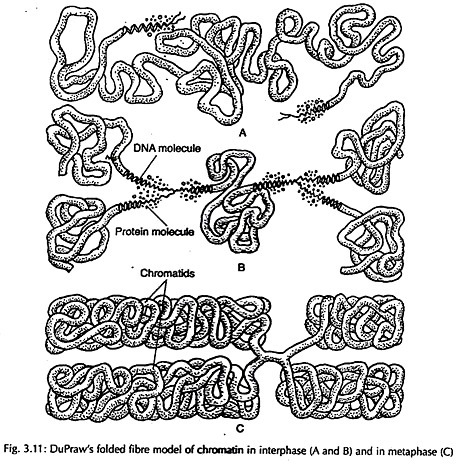
(i) Folded Fibre Model:
According to this model proposed by DuPraw, each chromatid of chromosome consists of a single fibre of 200- 500 Å in diameter folded both longitudinally and transversely (Fig 3.11).
(ii) Nucleosome model:
This is the most modern and acceptable model proposed by Roger Kornberg in 1974.
Nucleosome Concept:
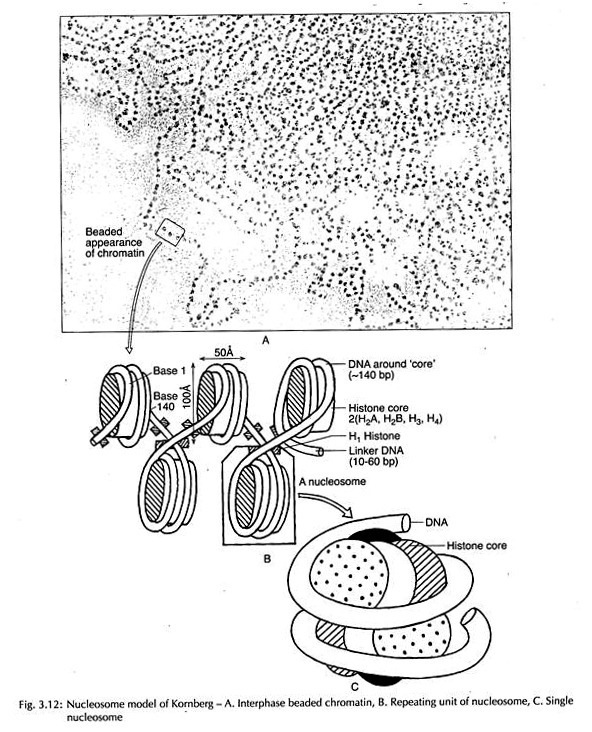
Electron microscopy and biochemical evidences have revealed that the chromosome (chromatin) structure is beaded in appearance (Fig. 3.12), each bead is termed nucleosome or Nu-body. Nucleosome is a complex of DNA and histone protein.
There are five histone fractions in the nucleosome assembly, of which four, i.e., H2A, H2B, H3 and H4 (two molecules each) enter into the composition of nucleosome and fifth one H, protein, serves as a linker between the nucleosome beads.
The octamer is surrounded by a DNA molecule having almost 200 bp of which approximately 140 bp enter into the coiling of octamers and the rest base pairs remain associated with the linker H1 molecule. The structure of nucleosome reveals evolutionary stability as noted even in yeast and human.
Experimental Evidence for Nucleosome: The evidences in support of nucleosome are as follows:
(a) X-ray diffraction pattern studies:
X-ray diffraction of chromatin indicated the presence of a structure repeating every 10 nm.
(b) Electron microscopic studies:
Electron microscopy of ruptured nuclei showed the presence of a series of spherical particles connected by a fine filaments the so-called beads on a string. The beads have a diameter of 7-10 nm but the length of the filaments is variable.
(c) Digestion of Chromatin with Micro-coccal Nuclease:
It is already stated that the nucleosomes grossly appear as beads on string. It is obvious that beads are connected by non-beaded string or linker DNA which holds the nucleosomes. When a small fragment of DNA containing 4-5 nucleosomes are treated with Micro-coccal nuclease, it gradually digests the linker DNA but nucleosomes remain partially resistant to nuclease action.
An analysis of the size of DNA showed that spacing between the successive nucleosomes was about 200 bp. On further digestion the size of mono-nucleosome with about 200 bp DNA is reduced first to 166 bp and finally to 146 bp and H1 is lost.
Association of Histones in Nucleosome:
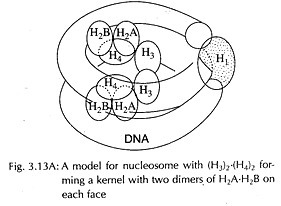
There are only five histones in each repeating unit. The four of them are H2A, H2B, H3 and H4 and the fifth one is H1. It was observed that H3 and H4 were present as a tetramer (a pair of dinners), i.e., (H3)2. (H4)2. it was shown that the chromosomes of most organisms had equal number of molecules of H2A, H2B, H3 and H4 and 25 nucleotides of DNA were present per histone molecule.
Since H3 and H4 are present as tetramers, each repeating unit may have one such tetramer and two molecules each of H2A and H2B. Histones will thus form an octamer (2H2A + 2H2B + 2H3 + 2H4) associated with 200 base-pairs in each repeating unit.
One molecule of H2 is associated with each repeating unit but absent in the core.
It was also suggested that tetramers make the core of the unit and oligomers determine the spacing thus giving a flexible structure. H2A and H2B were added as two dimers on each face of the tetramer (H3)2. (H4)2. The (H3)2. (H4)2 tetramer makes the central kernel associated with two independent dimers H2A- H2B (Fig 3.13A). DNA enters and leaves the same side and H, sealing off the two turns of DNA.
Coiling of DNA in Nucleosome:
On prolonged nuclease digestion, beyond the cleavage between nucleosomes, the DNA was removed from both the free ends yielding particles containing only 146 bp instead of 200 bp. This reduced form of nucleosome is called core- particle.
Adjacent core particles, each with core DNA, are joined with each other through DNA segment, called linker DNA which is removed during prolonged nuclease digestion. The core DNA has 146 bp but linker DNA can have 8 bp to 114 bp.
Three dimensional structure as revealed from x-ray crystallography shows that the nucleosome core-particle is neither spherical, nor perfectly flat, but wedge-shaped, about 11 nm in diameter and 6 nm high. The study of crystals suggested that DNA double-helix is coiled into a large helix or super helix by making two turns around the histones in the middle of the particle.
From the structure of DNA and diameter of core particle, it has been estimated that two super- helical turns would have 160 bp. Since the length of DNA in the core is only 146 bp, therefore it makes only 1¾ turns (Fig. 3.13B). The particle is, therefore, thin on one side accounting for the wedge-shape.
Modification of Nucleosome:
Nucleosome undergoes histone modifications and structural rearrangements to become transcriptionally active. Histone protein modification has been revealed by acetylation, phosphorylation, methylation and ubiquitination at specific amino acid residues in the histone tails affecting chromatin structure.
The core histones have two domains – histone fold domain involved in interactions with other histones as well as in wrapping DNA around nucleosome core, and an amino-terminal tail domain extending outside the nucleosome.
The amino-terminal tail domain of histone undergoes the modifications to increase the accessibility of chromatin to DNA-binding proteins for transcriptional activation (Fig. 3.14A):
(a) Acetylation at specific lysine residues;
(b) Phosphorylation of specific serine residues;
(c) Methylation of specific lysine and arginine residues; and
(d) Ubiquitination of specific lysine residues.
Combinations of specific histone modifications constitute a ‘histone code’ that regulates gene expression by recruiting other regulatory proteins to the chromatin template. Nucleosome structure is also, altered by nucleosome re-modelling factors facilitating the binding of transcription factors to chromatin by repositioning nucleosomes on the DNA through sliding of histone octamers along the DNA molecule (Fig. 3.14B).
Solenoid Model:
The 10 nm fibre of nucleosome gets coiled upon itself to form 30 nm wide helix with 5 to 6 nucleosomes per helix. In this helix successive turns come close together so that their centre to centre distance was about 10 nm. This 30 nm structure was called solenoid (Fig. 3.15).
The H1 proteins helped in folding of the 10 nm fibre into 30 nm solenoid because when H1 was removed this ordered folding was absent. H1 molecules aggregate by cross-linking to form polymers and may thus control the formation of solenoid.
The packaging of DNA in a chain of nucleosomes is about 5 to 7 times more compact than free DNA and in a 30 nm solenoid it is packed about 40-fold.
Loops, Domains and Scaffold:
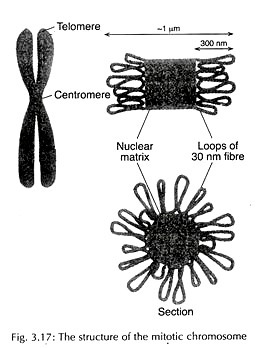
The 30 nm solenoid fibre is then organized into looped domain structure (Fig. 3.16). Each loop has approximately 50 turns of solenoid.
Each loop is with 85 kb of DNA and a length of 10-30 pm. These loops surround a central core of scaffold or matrix, made up of non-histone fibrous network of chromosomal proteins. This scaffold is involved in condensing solenoid fibre into tightly packed metaphase chromosome (Fig. 3.17).
The scaffold proteins include two abundant proteins – the larger DNA topoisomerase II and the smaller matrix protein. Both initiation and continued replication of DNA occur in association with matrix proteins and topoisomerase II binding sites are found on matrix associated DNA.
The binding sites for topoisomerase II are called Scaffold Attachment Regions (SAR) and matrix proteins bind Matrix Attachment Regions (MAR). It is believed that the same sequence of DNA works as MAR in interphase nuclei and as SAR in metaphase chromosomes.
Order of Organization:
Thus the chromosome fibre is presumed to be formed by three levels of coiling (Fig. 3.18):
(i) The first level of condensation involves packaging DNA as a supercoil into nucleosomes. This produces the 10 nm diameter interphase fibre.
(ii) Second level of condensation involves an additional folding and on supercoiling of 10 nm nucleosome to produce the 30 nm solenoid.
(iii) Finally solenoid fibre of 30 nm is organised into loops around a central scaffold to make the tightly packed metaphase chromosome.
Note # 6. Mechanism of Chromosome Condensation:
The interphase chromatin condenses approximately a thousand-fold to’ form the metaphase chromosome. Interactions between H1 histone molecules bound to the DNA as it enters each nucleosome core particle, are involved in the folding of chromatin into higher-order compact structure.
Histone H1 is a substrate for the Cdc2 protein kinase and is phosphorylated during mitosis. Phosphorylation of H3 histone for condensation of mitotic chromosome has also been indicated. Recent studies have identified a complex of five proteins called condensin, involved in condensation of interphase chromosome organization to metaphase chromosome.
This complex is composed of two structural subunits of SMC proteins and three regulatory subunits. Phosphorylation of regulatory subunits by Cdc2 kinase plays a major role in chromosome condensation. A similar complex cohesion plays a role in the pairing of sister chromatids at metaphase.
These proteins are long dimeric molecules hinged at the centre with a globular domain at each’ end that bind to DNA and hydrolyse ATP (Fig. 3.19). 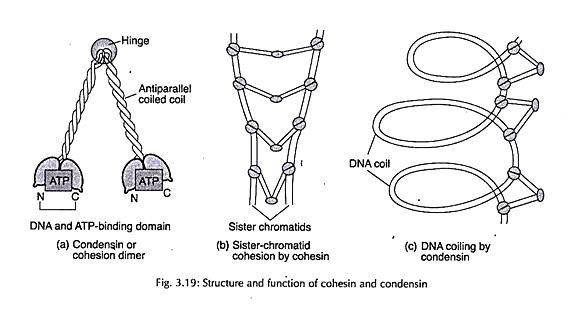
Note # 7. Special Types of Chromosomes:
i. Accessory (B) Chromosome:
Accessory chromosome otherwise termed as supernumerary chromosome or B chromosome, in its simplest form, can be defined as an extra chromosome that is not necessary for the individual and not homologous to any of the normal chromosomes. These chromosomes differ from the normal ones in their variable number, smaller size and greater degree of heterochromatinization.
They may vary in number within different tissues of the same individual, between different individuals of the same population and even between populations of the same species from different regions. They are usually aero- or telocentric in nature. Submetacentrics and metacentrics have also been recorded. They do not usually pair among themselves like normal chromosomes during meiosis.
Such chromosomes were first recorded by Longley and Randolph in maize in 1927-1928, who referred to them as B chromosomes as against the normal number of 2n=20 in a normal maize plant. These chromosomes were much smaller than the normal chromosomes (Fig. 3.20) and occurred in varying frequencies in different strains of maize.
In Allium stracheyii too, individuals differ in the number of B chromosomes, whereas absolute constancy of such accessory chromosomes in all individuals of a population has been recorded in Tradescantia virginiana.
In plants, the number of B chromosomes present may increase or decrease during the development of the germ line, particularly the archesporium and/or during gametophyte development. In Trillium, Lilium, Tradescantia and Plantago, univalent B chromosomes segregate preferentially to the functional megaspore in female meiosis.
In certain cases like Festuca difference in B chromosomes was observed in plants growing under different environmental conditions, e.g., clay soil and dry climates.
The centromere of B chromosome shows irregular behaviour resulting in irregular anaphasic separation and uneven segregation. The aberrant anaphasic separation or non-disjunction, which is responsible for the numerical variation of B chromosomes, may lead to variations within different populations of the same species such as in rye, Urginea indica, Poa alpina and Allium stracheyii.
Meiotic pairing of B chromosomes shows a wide range of variations in plants. It may be homologous between two similar B chromosomes, without chiasma formation or non-homologous in exceptional cases with A chromosomes such as in species of Clarkia. A typical pairing may involve an end to end association between A and B chromosomes as in Haplopappus spinulosus.
B chromosomes are usually heterochromatic, showing differential staining behaviour as in Tradescantia virginiana. Allium stracheyii and Secale cereale. The degree of heterochromatinization may range from the totally heterochromatic B chromosomes of Anthoxanthium to the totally euchromatic ones of Tradescantia. The amount of repetitive DNA seems to be involved as well.
The common property of accessory chromosomes is their deleterious action or in certain cases, their lack of activity. B chromosomes have been shown in certain cases to modify chromosome pairing in hybrids between closely related species, such as in Lolium, wheat and Festuca.
The origin of accessory chromosomes is rather obscure. They may have been derived from the ordinary chromosomes sometime in the remote past, starting possibly as heterochromatic centric fragments. The concept of B chromosome as being heterochromatic in nature has led to the understanding that these chromosomes are highly variable in structure and behaviour.
Their nature and the effects of their presence can be explained more plausibly taking into account the presence of repetitive DNA in large amounts in most of them, which in some cases, may be shared with the regular chromosomes.
ii. Lampbrush Chromosome:
The oocytes of some amphibians possess giant-sized chromosomes. These chromosomes can sometimes be seen even without a microscope. Their length is maximum during diplotene of meiotic prophase and in some cases may even reach 1000 pm.
During this stage each chromosome can be distinguished into a central heterochromatic axis and a large number of paired loops projecting from it. Because of their brush-like appearance at this stage, these chromosomes are known as ‘lampbrush chromosomes‘ (Fig. 3.21).
Autoradiography has revealed that the loops are sites of RNA synthesis, i.e., represent active chromatin. In oocytes, diplotene is of comparatively longer duration and during this period large quantities of messenger RNA are synthesized and stored for use during rapid protein synthesis at the time of embryo development. A DNA double helix forms the main axis of the loop.
The DNA of the main axis of chromosomes is inactive and, therefore, very much condensed, but the DNA of loops is active and stretched. When the activity ceases, loop DNA also condenses and, therefore, the loops disappear. Loop DNA appears thick because it is covered with non-histone proteins and nascent RNA molecules.
iii. Polytene Chromosome:
These also are examples of giant chromosomes. Most of the studies have been done with salivary gland chromosomes because these chromosomes are the largest. They arise due to many longitudinal splitting’s of chromatids and consequent non-separation of split portions. The process of chromosome duplication without cell division is called endomitosis.
The polytene chromosome of Drosophila are of special importance because they have transverse bands on them (Fig. 3.22). Dark staining bands alternate with lightly staining inter-band regions of chromosomes. Many polytene chromosomes show various types of swellings or puffs (Balbiani rings) in their inter-band or band regions, specially during growth and differentiation of larvae.
Polytene chromosomes in plants arising out of endomitosis have also been recorded in endosperm, haustoria and suspensor cells.
iv. Sex Chromosome:
In plant system where the male and female plants are distinct from each other, the chromosomes associated with sex can be located. However, only a few species (Melandrium, Coccinia) have been shown to have distinct sex chromosomes (Fig. 3.23). Details of sex chromosomes and sex determination have been dealt with in another chapter.
However, the most common sex determining mechanism is XY, where X and Y chromosomes differ from each other. The female plant is represented by two sex chromosomes XX, in addition to autosomes. Male is represented by a heteromorphic pair XY, forming a bivalent. Both the chromosomes show hetero-chromaticity, Y being more heterochromatic than X-chromosome.
Their segregation during meiosis is normal. In the female, all the gametes contain X-chromosomes whereas in male, half the gametes contain X-chromosomes and half the gametes Y-chromo- some. Sex chromosomes in general are late replicating, compared to autosomes and they can be differentiated from autosomes by staining behaviour.
Note # 8. Karyotype Concept of Chromosome:
Karyotype concept was developed by S. Navashin based on the observations that the number of chromosomes and morphology of each chromosome pair is normally constant and characteristic for a species. The term ‘karyotype’ is used for a group of characteristics that allow the identification of a particular chromosomal set.
It is the phenotypic appearance of the entire chromosome complement of the species representing all the chromosome types based on their morphology.
Karyotype study helps to represent the origin and evolutionary relationship among the different taxa. Depending on the differences between smallest and largest chromosome of the set, the karyotype may be symmetric (less difference) or asymmetric (large difference). Increased karyotype asymmetry is associated with advanced group of plants.
Parameters: The parameters used in karyotype preparation are:
(i) The number of chromosomes,
(ii) The length of each chromosome,
(iii) The length of chromosome arms (both short and long arms),
(iv) The position of centromere,
(v) The existence and localization of secondary constriction,
(vi) The position and size of the satellites,
(vii) Absolute and relative chromosomal size,
(viii) Basic chromosome number,
(ix) The degree and distribution of heteromatic regions,
(x) Amount and location of repeated sequence.
Idiogram is the diagrammatic representation of karyotype showing all the morphological features of the chromosomes grouped on the basis of position of centromere and ordered in a series of decreasing size. Karyogram is the actual representation of the karyotype performed from the microphotograph or drawing (Fig. 3.9).
Centromeric index (‘i’ value) is calculated to determine the location of centromere on the chromosome.
Note # 9. Heterochromatin in Chromosome:
Chromosomes are principally composed of two components – heterochromatin and euchromatin. Euchromatin constitutes the principal functional regions of chromosomes. Heterochromatin on the other hand, is present on both sides of the centromere, at the telomeres, secondary constrictions as well as at the intercalary segments (Fig. 3.10).
The term ‘heterochromatin’ was originally derived from heterochromosome or sex chromosome, differentiating it from autosomes by Heitz. Since sex chromosomes differ from rest of the chromosomes in their staining behaviour, the chromosome segments showing staining cycle, similar to sex chromosomes, were called heterochromatic.
However, Darlington attributed to heterochromatin, the property of allocycly. It implies, positive staining in chromosomes or heteropycnosis during interphase and reverse behaviour in metaphase. However, in a number of cases, the chromosomes may be apparently condensed or de-condensed throughout, as in the secondary constriction region.
As such, all these segments were clubbed under the category of heterochromatin, the common property being staining behaviour different from rest of the chromosome segments.
According to Vanderlyn (1949); any segment displaying properties different from those of euchromatin should be termed heterochromatin. This statement does not necessarily imply that all heterochromatic regions, including those of primary and secondary constriction regions are fundamentally alike.
Heterochromatin thus covers a rather heterogenous assemblage of chromatin represented by diverse characteristics in different species. The terms applied to heterochromatic regions earlier were constitutive or inherited and facultative or appearing during development.
The best example of facultative heterochromatin manifested during development is provided by mealy bug, where one set of chromosome becomes heterochromatized during development and other remains euchromatic. In sex chromosomes of mammals, due to dosage compensation, a single X remains active, the other becoming inactive or heterochromatized.
In plants, following cold treatment, certain regions of chromosomes often show negative staining as in species of Fritillaria, Trillium and Paris. Such areas, termed by Darlington as nucleic acid starved areas arising out of environment changes, come also under the category of facultative heterochromatin.
The constitutive heterochromatin, on the other hand, remains stable in the chromosome and appears condensed during interphase as noted in a large number of plant species including Vicia faba. It is also characterized by late or early replication of DNA, different from that of euchromatin, as in case of sex chromosomes.
The best example of constitutive heterochromatin is provided by segments on both sides of the centromere which remains as single condensed block during metaphase being designated as prochromosome.
In addition to centromere as discussed above, constitutive heterochromatin is also located at the telomeric regions, nucleolar organizing regions as well as intercalary positions in certain organisms (Fig. 3.10). The entire chromosome may also be heterochromatic, such as sex chromosome in some plants and accessory chromosomes.
It was also presumed that genetically heterochromatin, though representing condensed state of chromatin, may be made up of genes having small, similar and supplementary effects, as embodied in the concept of polygenes of Mather.
In later years, the discovery of repetitive DNA sequences and their distribution in chromosome indicated that heterochromatic segments are made up of repetitive DNA sequences, both GC/AT-rich, the number and condensed state varying in different segments of chromosomes.
Some segments like pro-chromosomes show high homogeneous repeats. No specific qualitative functions have been attributed so far to heterochromatin. However, such segments may be involved in regulating chromosome metabolism, cytoplasmic synthesis and non-specific functions at the chromosomal level which are all categorized as nucleotypic.
Its variations at different taxonomic level make it suitable for the study of biodiversity. Genotypes may differ in the nature of heterochromatin.
Regarding the functions, constitutive heterochromatin helps in pairing of homologues as studied in Triticale. It also influences the shortening of the duration of cell cycle as noted in several species growing in stress situations like desert and alpine regions. During evolution, moreover, there is often a change in total DNA amount to great extent, which principally confined to the repeated DNA sequences of heterochromatin.