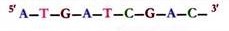This article provides a brief case study on recent trends in animal biotechnology.
Contents
Introduction:
Animal biotechnology has been defined in various ways. One of the most recent quoted definitions of animal biotechnology is: “The application of scientific and engineering principles to the processing or production of materials by animals or aquatic species to provide goods and services”.
Animal Biotechnology has developed rapidly since early 1980s when the first transgenic mice and first in vitro produced bovine embryos were created.
An attempt has been made in the present article to describe the information sources and processes which are available in the field of animal biotechnology.
Examples of animal biotechnology include generation of transgenic animals or transgenic fish using gene knockout technology to generate animals in which a specific gene has been inactivated, production of nearly identical animals by somatic cell nuclear transfer, or production of infertile aquatic species.
To cater to the need of our fast growing population, searching cheap sources of proteins either from animals or from plants have been the most daunting task for us. Animal protein is an excellent source of dietary amino acids essential for human nutrition. In addition, we are able to harvest meat, milk and fibre from live animals and use them for different welfare programmes. The production of food from animals is expensive and less efficient than plants. In spite of these disadvantages, we continue to farm animals on commercial basis for food, fibres and by-products.
Our ancestors first domesticated animals many thousands of years ago and since then they have been our constant companions. From that time onwards, the animals themselves started to change. With the passage of time and continued isolation from their wild relatives, domestic animals started to display different characteristics. Some would have produced more or better milk than others, some would have grown faster or been more fecund, some would have been more vigorous.
Our ancestors might .have noticed these variations and mated animals with similar desirable traits, but they did so without understanding the basis of heredity. Restrictive breeding and selective mating gradually led to the many diverse breeds of livestock we have today, although most breeds of the dominant domestic cattle, Bos taurus, arose in the last couple of centuries. Herds of ancestral species, Bos primigenius, lived in Europe in the 18th century.
Thus animal improvement is no longer a matter of interest but it is a matter of necessity. Biotechnology provides us powerful tools that engage us learning the basics of these tools for increase in animal production world wide. In this article I will discuss what these tools are and their applications.
A. Reproduction in Animals/Reproductive Technology:
Reproduction has some important roles in production system. It must occur at a rate that ensures replenishment of stock is greater than its use and improving genetic quality of stock. The aim of reproductive technologies is to increase the number of progeny while improving the genetic quality of stock in general and most intensively managed animals like dairy cattle and domestic pigs in particular. This has been done by various procedures like artificial insemination, embryo transfer, in vitro fertilization and embryo cloning.
A. 1. Artificial Insemination (AI):
Artificial insemination, the first animal biotechnology, takes advantages of the male’s excess gametes (sperm) production capacity. This has been the single-most important factor in increasing the productivity. There are many variations of artificial insemination to consider. There is animal-animal artificial insemination, fresh-extended (chilled) semen insemination and frozen semen insemination.
Before insemination the sperm ejaculated is collected, diluted and examined under microscope to determine the number and mortality of sperms. Techniques of AI include vaginal deposition, surgical implant, and trans-cervical insemination. Studies have been published with convincing evidence that every semen preparation in fresh, fresh-extended or frozen will produce larger litters if semen is deposited into the uterus, especially with trans-cervical technique. Fresh and fresh-extended semen produce good results with vaginal deposition.
For cattle, AI is done in standing animals without using anesthesia, using a procedure known as “rectal palpation”. Of the different types of semen preparation, it is again obvious that fresh animal-animal collection and insemination will give the best semen preparation. Fresh-extended or “chilled” semen has performed very well for many years, but not that well as does the fresh semen.
The worst results are with the frozen semen, no matter what process is used for the freezing. Females are inseminated after ovulation, which last only a few hours and occurs most commonly at night. In many breeds of farm animals it is very difficult to detect ovulation (oestrous).
The alternative is to induce females to ovulate in a synchronized manner. In practice, it is rather impossible to achieve total synchrony of ovulation but about 80% of females would respond to the inducer. With ruminants, ovulation in females can be induced by administration of hormone like progesterone or prostaglandin.
Single sex is being preferred by all livestock industries. In dairy and beef cattle, females are more accepted than males because females are not efficient in reproduction and having desirable production traits. Same thing happens in pig industry. The sex of the embryo is determined solely by sperm containing either X chromosome or Y chromosome.
Since half of the sperm in an ejaculate contain the X and the remaining half the Y, the sex ratio of progeny is near to 1 : 1. Separating sperm containing the X chromosome from that of the Y would allow us to manipulate the sex progeny by AI. Separation of the two populations has been done using a fluorescent activated cell sorter (FACS). The method is very expensive and very slow too. Monoclonal antibody that binds specifically to Y-bearing sperm cells could be used in future to separate Y- bearing and X-bearing sperm. The flow sorting of sperm cells to separate X from Y bearing cells has been successful in most cases tested and in cattle and swine has resulted in offspring of the desired sex.
A. 2. Embryo Transfer:
The propagation of genetically valuable animals is enhanced by transferring embryo. To do this one has to increase the number of mature eggs produced by a selected female. Following fertilization, those fertilized eggs are implanted into foster mother, one for each egg. Donor females are injected with prostaglandin F2a (PGF2a) to induce a synchronized oestrous before treatment is started. Ten days after oestrous period they are injected with FSH for a period of four days, followed by PGF2a to induce oestrous.
Superovulated donors are then mated by artificial insemination. Six to eight days after insemination, the fertilized eggs are recovered from cattle and six days from sheep and goat. They are immediately transferred into synchronized recipients. Alternatively, they can be frozen for indefinite storage. With good practice, pregnancy rates of 50-60% can be achieved in cattle from transferred embryo.
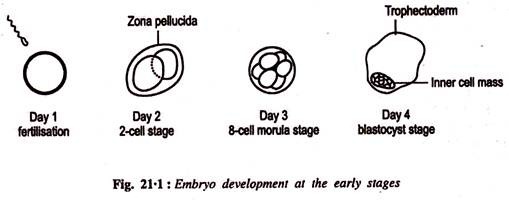
Pregnancy output rates can be increased by embryo splitting; a method which produces identical twins. Recently this has become a routine production tool, requiring micromanipulation equipment and minimal training. Embryos were obtained from donor at the blastocyst stage (Fig. 21.1) and transferred into a standard cell culture medium containing hypertonic sucrose and bovine serum albumin.
The embryo is then transferred to a plastic Petridish containing standard culture medium, where it sinks to the bottom and sticks to it. It occurs because of the electrostatic interactions between the BSA-induced negative charge on the outer membrane and the positive charge of the plastic disc.
The embryo is then bisected using a micromanipulator fitted with a fine blade under the inverted microscope. During embryo splitting care must be taken to ensure that the inner cell mass is split into equal halves. After splitting, they are transferred into the oviduct of the recipient female following the same procedure as for normal transfer. Routine biopsy has been carried out to determine the sex of progeny, a production trait. Presence of Y chromosomal DNA in cells of an embryo biopsy indicates that the embryo is a male: if no Y chromosomal DNA is present it is female. This is being done using PCR to amplify Y-chromosome specific DNA.
A. 3 In Vitro Fertilization (IVF):
Embryo transfer is not used widely because of many factors including high costs, technical difficulty and limited availability. We can overcome these problems if oocytes, collected from a donor female are fertilized in vitro. It would ensure fertilization of more eggs using less amount of semen. Thus IVF is more efficient method than AI. During oestrous cycle a number of Graafian follicles start growing.
Eventually one of the follicle matures and ruptures, releasing the eggs for fertilization. Oocytes are recovered from these follicles from super ovulated donors by laparoscopic surgery. Eggs are then incubated to be matured and fertilized in vitro. This method demands high technical skill and, therefore, not always cost effective.
The success of IVF depends on collection of large number of oocytes and there maturation in vitro. This can be done by removing an ovary from a selected female and is induced to mature in vitro. Numbers of scientists across the globe have been carrying out research on oocyte maturation in vitro. However, it may be worthwhile to mention here that majority of Graafian follicles never attain maturity. The highest success rates with embryo transfer are reported when blastocysts are implanted into recipients.
This can be done if embryos are maintained in culture. Many laboratories have shown that 60% of IVF cattle embryos can be cultured to blastocyst stage. High rate of abortion is recorded from culture embryos during the first two months of pregnancy. This may be due to genetic defect in the oocyte and or fertilizing sperm and environmental mutagenesis of egg, sperm or embryo (mainly caused by oxygen free radicals).
B. Cloning:
Another application of animal biotechnology is the use of somatic nuclear transfer to produce multiple copies of animals that are merely identical copies of other animals. This process is called ‘cloning’. So cloning is the production of one or more identical plants or animals that are genetically identical to another plant or animal.
It would facilitate increased production of numbers of one particular embryo having desirable characteristics. Nature itself is the greatest cloning agent. In about one of every 75 conception, the fertilized embryo splits and produces monozygotic twins.
B. 1. DNA Cloning:
To do further manipulations and analysis of individual recombinant DNA molecules (e.g. DNA containing insulin gene), we must have many copies of the molecules, usually in a purified form. DNA cloning is a technique to produce large quantities of specific DNA segment. The DNA segment to be cloned is inserted to a vector, which is a vehicle for carrying inserted DNA into a suitable host cell, such as the bacterium E. coli.
The vector contains sequences that allow it to be replicated within the host cell and usually refer to as cloning vector. There are numerous cloning vectors in current use and choice among them depends on the size of the DNA fragment that needs to be cloned or the use to which the clone will be put.
B. 1a. Plasmid Vectors:
Bacterial plasmids are small circular DNA molecules that are distinct from the main bacterial chromosome. They replicate their DNA independently of the bacterial chromosome. The plasmids that are routinely used as vectors are those that carry genes for drug resistance. The drug resistance genes are useful because drug- resistant phenotype can be used to select for those cells that contains the recombinant plasmid.
The process by which bacterial cells take up DNA from the medium is called transformation. This forms the basis for cloning plasmid in bacterial cells (Fig. 21.2). In most commonly used methods recombinant plasmids are added to a bacterial culture that has been pretreated with Calcium ions. Bacterial cells are then activated (by heat-shock) to take up DNA from the medium.
Normally a small number of cells are able to take up and retain one of the recombinant plasmid molecules. Once a bacterial cell has taken up a recombinant plasmid from the medium, the cell gives rise to a colony of cells containing the recombinant DNA molecule. These bacteria containing a recombinant plasmid are selected from the rest by growing the cells in the presence of the antibiotic specific to drug-resistance gene of plasmids.
B. 1b. Bacteriophage Vectors:
There are different classes of bacteriophage vectors, depending on whether chromosomal DNA inside the bacteriophage is single stranded or double stranded and the size of the donor DNA insert.
B. 1c. Bacteriophage (Lambda):
It is used as a cloning vector for double-stranded DNA inserts up to approximately 25 kb. Lambda phage heads harbor a linear DNA (genome) of about 50kb in length. The central part of the genome is not required for replication or packaging and so a central part can be cut out by using restriction enzyme and discarded.
The two remaining ‘arms’ at either end of the genome are ligated to restriction-digested donor DNA (Fig. 21.3). The recombinant molecules can be introduced into E. coli by transformation. Once in the bacteria, the donor DNA insert is amplified along with the lambda/phage DNA and packaged into a new generation of virus particles, which are released when the cell is lysed. The released particles infect new cells. This results into the occurrence of a clear spot (or plaque) in the bacterial plate at the site of infection. Each plaque harbors millions of large particles, each carrying a single copy of same donor DNA insert.
B. 1d. Vectors for Larger DNA Inserts:
The maximum size of the donor DNA that can be inserted into a standard plasmid or vector is about 30 kb in length. To meet these demand, several vectors have been engineered. The largest prokaryotic inserts use the BAC (bacterial artificial chromosome) vector system. It is based on the 7-kb F plasmid and has the ability to accept larger DNA inserts (up to about 300 kb).
For inserts larger than 300 kb, YAC (yeast artificial chromosome) — a eukaryotic vector system based on yeast chromosomes introduced into yeast cells by transformation, is being used to clone recombinant molecules as large as 1,000 kb in length.
B. 1e. Formation of a DNA Library:
DNA cloning is frequently used to produce DNA libraries, which are collections of cloned DNA fragments. There exists two basic types of DNA libraries: genomic libraries and cDNA libraries. Genomic libraries are produced from total DNA obtained from nuclei and contain all of the DNA sequences of the species.
Once a genomic library of a species is created, scientists can use the library to isolate specific DNA segments. cDNA libraries are derived from DNA copies of mRNA population. cDNA libraries are produced from mRNA present in a particular cell type and this corresponds to the genes that are active in that type of cell.
B. 1f. Identification of DNA Molecules of Interest:
Immediate after cloning, the next task is to find that particular clone which contains the desired gene. This has been done either by using a specific probes (for finding DNA or protein) or by probing a specific nucleic acid.
B. 1g. Use of Recombinant DNA Technology for Genetic Engineering:
The use of sophisticated recombinant DNA techniques to alter the genotype and phenotype of an organism is called genetic engineering. This is being done by introducing genes into eukaryotic cells, where they are transcribed and translated. There are number of strategies to achieve this. The most frequently used technique is the viral-mediated gene transfer, which is termed transduction.
Here the engineered DNA is incorporated into the genome of a non-replicating virus and allow the virus to infect the cell. It is the type of virus that would determine whether gene of interest can be expressed temporarily or integrated into the genome of the host cells. Retroviruses have been used in gene therapy to transfer a normal gene into the cells of a patient having a defective gene. Transfection is a process by which naked DNA can be introduced into cultured cells.
During the process the cells are treated with either calcium phosphate or DEAE-dextran, both of which form a complex with the added DNA that promotes its adherence to the cell. It is observed that only a few cells take up the DNA and incorporates it stably into the chromosomes to make a transgenic eukaryote (Fig. 21.4). Electroporation and lipofection—the two other methods have also been used to transfect cells.
In lipofection, foreign DNA binds to positively charged lipids (liposomes) which are capable of fusing with lipid bilayer of the cell membrane and supplying the DNA to the cytoplasm. In electroporation, foreign DNA finds their way through the modified plasma membrane, induced by electric current, into the nucleus and become integrated into the genome. Foreign DNA can also be introduced into a cell by microinjecting directly into the cell nucleus.
For a long time Xenopus oocytes have been used to study the expression of foreign genes. It contains all ingredients for mRNA synthesis. When foreign DNA is delivered into the nucleus, it initiates transcription and eventually m-RNA is transported to the cytoplasm, where they are translated into proteins that can be detected immunologically.
Another important target for injected DNA is the nucleus of a mouse embryo. Here the foreign DNA become integrated into the egg’s chromosome, which will pass on to all the cells of the embryo and finally to the adult. Those animals that have been genetically engineered are called transgenic animals.
The first transgenic animal was created by Ralph Brinster of the University of Pennsylvania and Richar Palmiter of the University of Washington in the year 1981. They succeeded in introducing a gene for rat growth hormone (GH) into the fertilized eggs of mice. The injected DNA was constructed in such a way that the rat GH gene (coding protein) is located downstream from the promoter region of the mouse metallothionein gene.
In a normal rat, the synthesis of metallothionein gene is accelerated following the treatments of metals like cadmium, zinc, or glucocorticoid hormones. In the transgenic mice, synthesis of the GH gene has seen to have enhanced following treatment with metals and glucocorticoids. Mice are the most important models for mammalian genetics. The technology developed in mice is applicable to human.
There are two strategies for transgenesis in mice: ectopic insertion and gene targeting. The procedure involved in ectopic insertion is simply to inject bacterially cloned DNA solutions into the nucleus of early-stage embryos, which has been discussed earlier. Gene targeting is a bit rare event and a multi-step process is needed involving the use of embryonic stem cells. Embryonic stem cells (ES) have the ability to form any and all parts of a mouse, that is why they are called totipotent cells.
The process of gene targeting can be recognized by one of its outputs, the substitution of a nonfunctional gene for the normal gene. This type of targeted inactivation is called a gene knockout. Here embryonic stem cells are isolated from a mouse strain (albino) and embryos are transfected with a DNA insert containing an inactive, mutant allele of the gene to be knocked out.
The ES cells are then injected into the recipient young mouse embryo (blastocoel) collected from an albino strain. In the next step, the embryo containing the ES cells grows to term in surrogate mother. The resulting progeny are chimeric, having tissue derived from both the donor (transplanted ES) and recipient strains. Chimeric mice are then mated with their sibling to produce homozygous mice with the knock-out in each copy of the gene (Fig. 21.5). These are knock-out mice that lack a functional copy of the gene.
The term ‘cloning’ usually refer to three different procedures. The three types of cloning are: embryo cloning, adult DNA cloning, and therapeutic cloning.
B. 2. Embryo Cloning:
It might be more accurately called ‘artificial twining’ because it stimulates the mechanism by which twins naturally develop. It involves removing one or more cells from an embryo and encouraging a cell to develop into a separate embryo with the same DNA as the original.
It has been successfully carried out for years on many species of animals including cattle, sheep, pigs, goats, horses, moles, cats, rats and mice. The technology of embryo cloning has been done in two ways: nuclear transfer and embryonic stem cell. Cloning of embryos has been used in mice since the late 1970s and animal breeding since 1980s.
B. 2a. Nuclear Transfer:
The technique involves culturing somatic cells from an appropriate tissue (preferably fibroblast) from the animals to be cloned. Nuclei from the cultured somatic cells are then micro-injected into an enucleated oocyte obtained from another individual in same or closely related species.
Through a process that is not yet understood, the nucleus from the somatic cells is reprogrammed to a pattern of gene expression suitable for directing normal development of the embryo. After further culture and development in vitro, the embryos are transferred to a recipient female, and, ultimately, would result in the birth of live offspring.
The success rate of producing animals by nuclear transfer is less than 10% and depends on many factors including the species involve, source of recipient ova, cell type of donor nuclei, treatment of donor cells prior to nuclei transfer and techniques used for nuclear transfer.
The first publicly announced human cloning was done by Robert J. Stillman and his team at the George Washington Medical Centre in Washington D.C. They took 17 genetically defective human embryos. These embryos were derived from ovum that had been fertilized by two sperms. This has resulted into an extra set of chromosomes which ruined the ovum’s fate. None could have developed into a fetus. These ovum were successfully split in 1994, each producing one or more clones.
B. 3. Adult DNA Cloning (Reproductive Cloning):
This technique is used to produce a duplicate of an existing animal. It has been used to clone a sheep and other mammals. Reproductive cloning was earlier thought to be impossible in all mammals until it was activated in 1996 by a scientist, Dr. Ian Wialmut of the Roslin Institute in Roslin, Scotland, U. K. “Dolly”, a seven-month old sheep, was displayed to the media on February 23rd, 1997. She is the first large cloned animal using DNA from another adult.
A cell was taken from the mammary tissue of a mature six-year old sheep while its DNA was in dormant state. It was fused with sheep ovum which had its nucleus removed. The ‘fertilized’ cell was then stimulated with an electrical pulse. Out of 277 attempts at cell fusion, only 29 began to divide. These were all implanted in ewes, 13 became pregnant but only one lamb, Dolly, was borne. On March 4, 1997, President Clinton ordered a widespread ban on the Federal funding on human cloning in the USA. Research on human cloning continues in other countries.
Over the past 25 years tremendous advances have been made in the analysis of eukaryotic genomes. This progress began as molecular biologists learned to construct recombinant DNA molecules, which are molecules containing DNA sequences derived from more than one source.
It involves the isolation from the genome of a particular segment of DNA that codes for a particular polypeptide, isolation of enzymes which would cut DNA at precisely defined locations (restriction endonucleases) and those which would covalently join DNA fragments (ligases).
The Main Steps of DNA Cloning are given in (Fig. 21.6) and Involve the Following Procedures:
1. Isolation of the DNA to be cloned.
2. Insertion of the isolated DNA into another piece Of DNA called a vector, which is a vehicle for carrying foreign DNA into suitable host cell such as the bacterium E. coli. Although (Fig. 21.2). shows the use of a plasmid vector, other types of vector, can also be used such as the Bacteriophage lambda.
3. Transfer of the recombinant vectors in bacterial cells either by transformations by infection using viruses.
4. Selection of those cells which contain the desired recombinant vectors.
5. Growth of the bacteria to give as much cloned DNA as is needed.
B. 3a. Isolation of DNA to be Cloned:
The organism under study, which will be used to donate DNA for the analysis, is called the donor organism. Three sources of DNA segment can be considered: genomic DNA, complementary DNA and chemically synthesized DNA.
Genomic DNA:
These DNA sequences exist in the chromosomes of the organism under study and thus are the most easy source of DNA.
Complementary DNA (cDNA):
Complementary DNA or cDNA, is essentially a double-stranded DNA version of an mRNA molecule. cDNA is made from mRNA with the use of a special enzyme called reverse transcriptase, originally isolated from retroviruses. With the use of an mRNA molecule as a template, reverse transcriptase synthesizes a single-stranded DNA molecule that can be used as a template for double- stranded DNA synthesis. Synthesis of cDNA is very important in the analysis of gene structure and gene expression (Fig. 21.6).
Chemically Synthesized DNA:
Techniques for the chemical synthesis of oligonucleotides have been developed to produce DNA sequence for the purpose of recombinant DNA.
B. 3b. Construction of Recombinant DNA:
Isolated DNA molecules are broken into small fragments by enzymatically digesting it with endonucleases (restriction enzymes) that cleave at specific sites. Restriction enzymes cut at specific DNA target sequences (involving 4 to 8 nucleotides) and this property is one of the key features that make them suitable for DNA manipulation.
It is purely, by chance, any DNA molecule, be it derived from viruses, bacteria, plant and animal, contains restriction enzyme target sites. Thus, in the presence of appropriate restriction enzyme, the DNA would be cut into a set of small fragments according to the location of the restriction sites.
The Restriction Enzyme EcoR1 (from E. coli) Recognizes the following 6-Nucleotide-Pair Sequence in the DNA of Any Organism :
5′-GAATTC-3′
3′-CTTAAG-5′
This type of segment is called a DNA palindrome, which means that both strands have the same nucleotide sequence but in antiparallel orientation. EcoR1 recognizes and cuts only within the GAATTC palindrome sequence.
The Cuts are Between The G and The A Nucleotides on Each Strand of the Palindrome:
5′-G-AATTC-3′
3′-CTTAAG-5′
This staggered cuts leave a pair of identical five-base long single stranded ‘sticky ends’. The ends are called sticky because, being single stranded, they can be hybridized through base-pair hydrogen bonding to a complementary copy. The production of this sticky ends is another feature of many restriction enzymes that makes them suitable tools for recombinant DNA technology.
If two DNA molecules are cut with the same sticky end-producing restriction enzyme, the fragments of each will have the same complementary sticky ends, enabling them to hybridize with each other under the appropriate conditions in a test tube. (Fig. 21.7). illustrates the restriction enzyme making a single cut in the circular DNA molecule such as a plasmid (vector); the cut opens up the circle, and the resulting a liner molecule has two sticky ends.
If such a molecule is mixed with a different DNA molecule (donor DNA fragment containing insulin—gene cut with EcoR1, as shown in (Fig. 21.7), the two can then hybridize to each other through complementary sticky- ends to form a recombinant molecule. There are two reasons why the donor DNA containing insulin gene must be attached to plasmid DNA segments to form useful recombinant molecules.
First, a fragmented donor DNA segment does not have a necessary DNA sequences, on its own, to enable it to be replicated in a test tube or inside a host organism. The donor DNA must be physically attached to other DNA segments that can support replication in a test tube or inside a host cell. Second, an experiment may demand that multiple fragments be glued together to form a functional unit (e.g. a transcriptionally active gene).
Most commonly, both donor and plasmid DNA are digested together in the presence of DNA ligase. During the incubation, two types of DNAs become hydrogen bonded to one another by their sticky ends and then ligated to form circular DNA recombinants.
B. 4. Therapeutic Cloning:
This is a procedure whose initial stages are identical to adult DNA cloning. However, the stem cells are removed from the pre-embryo with the interest of producing tissue or a whole organ for transplant back into the person who supplied the DNA. The pre-embryo dies in the process. The goal of therapeutic cloning is to produce a health copy of a sick person’s tissue or organ for transplant.
The tissue or organ, would have the sick persons original DNA and, therefore, the patient need not have to take immunosuppressive drugs for the rest of life. Scientists are attempting to create transgenic pigs which have human genes. Their heart, liver or kidneys might be used as organ transplants in humans. This will save many lives because thousand of people die each year waiting for human organs.
Once achieved, transgenic animals could be cloned to produce as many organs as are needed. Researchers have produced transgenic animals typically in order to produce human hormones or proteins in its milk. These substances can be separated from the milk and be used for treating human ailments.
B. 4a. Gene Therapy:
Gene therapy is a technique for correcting genes responsible for disease development. The promise of gene therapy was evident from the first trials initiated in 1989. The disease addressed was adenosine deaminase (ADA) deficiency, a rare autosomal recessive disorder in which the body’s lack of a housekeeping enzyme is harmful in the immune system.
There, the absence of ADA results in premature death of T lymphocytes, leading to an immunodeficiency state (SCID – severe combined immunodeficiency). At the National Institute of Health, USA investigators transferred a normal ADA gene into the lymphocytes of children born with the disorder.
In later trials, the gene was put into bone marrow cells. In both strategies, the manipulation was ex vivo. Patient was injected with her own blood cells transfected with an wild type ADA gene. Parallely she was given adenosin deaminase obtained from cattle blood. Although long-term clinical benefits have yet to be appreciated, these early efforts served to establish the potential of gene delivery as a therapeutic strategy (Fig. 21.8).
Researchers do Use One of Several Approaches for Correcting Defective Genes:
a. A normal gene may be inserted into a nonspecific location within the genome to replace a nonfunctional gene. This approach is most commonly practised.
b. An abnormal gene could be swapped for a normal gene through homologous recombination.
c. The abnormal gene could be repaired through selective reverse mutation, which returns the gene to its normal function.
d. The regulation (the degree to which a gene is turned on or off) of a particular gene could be altered.
In most gene therapy studies, a “normal” gene is inserted into the genome to replace an “abnormal”, disease-causing gene. A vector must be used to deliver the therapeutic gene to the patient’s target cells. Currently, the most common vector is a virus that has been genetically altered to carry normal human DNA. Viruses have evolved a way of encapsulating and delivering their gene to human cells in a pathogenic manner. Scientists have tried to advantage this capability and manipulate the virus genome to remove disease-causing genes and insert therapeutic genes.
Target cells such as the patient’s liver or lung cells are infected with the viral vector. The vector then unloads its genetic material containing the therapeutic human gene into
the target cell. The generation of a functional protein product from the therapeutic gene restores the target cell to a normal state.
There are Number of Gene Therapy Vectors. Some of the Vectors are:
Retroviruses:
A class of viruses that can create double-stranded DNA copies of their RNA genome. These copies of its genome can be integrated into the chromosomes of host cells. Human immunodeficiency virus (HIV) is a retrovirus.
Adenoviruses:
A class of viruses with double-stranded DNA genome that cause respiratory, intestinal, and eye infections in humans. The virus that causes the common cold is an adenovirus.
Adeno-Associated Viruses:
A class of small, single-stranded DNA viruses that can insert their genetic material at a specific site on chromosome 19.
Herpes Simplex Viruses:
A class of double-stranded DNA viruses that infect a particular cell type, neurons, Herpes simplex virus type is a common human pathogen that causes cold sores.
Nonviral Vectors:
A gene is given a more or less artificial carrier; in some cases, a lipid encapsulation, so as to facilitate passage across cell membrane; in others, a tiny gold bead on which the gene is plated so it can be shot into the skin; in still others, a protein to which the gene is attached.
Requirements of Gene Therapy:
The gene (for example ADA gene) is identified and cloned. It is then inserted into patients own cells (ex vivo), treating them in tissue culture and returning them to the patient. It must be inserted into DNA, transcribed and translated with sufficient efficiency so that worthwhile amount of the enzyme is produced.
Retroviruses are used as the gene vector. Retroviruses have several advantages for introducing genes into human cells – their envelop protein enables the virus to infect human cells. RNA copies of the human ADA gene can be incorporated into the retroviral genome using a packaging cell.
Packaging cells express (i) an RNA copy of the human ADA gene along with a packaging signal (P) needed for the assembly of fresh virus particles and inverted repeats (R) at each end to aid insertion of the DNA copies into the DNA of the target cells,
(ii) an RNA copy of the retroviral gag, pol and env genes but with no packaging signal so that then genes cannot be incorporated in fresh viral particles.
Treated with these Two Genomes, the Packaging Cell Produces a Crop of Retroviruses with:
(i) The envelope protein needed to infect the human target cell,
(ii) An RNA copy of the human ADA gene, complete with R sequences at each end
(iii) Reverse transcriptase needed to make a copy of the ADA gene that can be inserted into the DNA of the target cell,
(iv) None of the genes (gag, pol, env) that would enable the virus to replicate in its new host. Once the virus has infected the target cells, this RNA is reverse transcribed into DNA and inserted into the chromosomal DNA of the host.
Besides, there are several other options for gene delivery. The simplest method is the direct introduction of therapeutic DNA into target cells. This approach is limited in its application because it can be used only with certain tissues and requires large amount of DNA.
Current Status of Gene Therapy Research:
The Food and DRUG Administration (FDA) has not yet approved any human gene therapy for sale. Current gene therapy is experimental and has not proven very successful in clinical trials. Little progress has been made since the first gene therapy clinical trial began in 1990.
In 1999, gene therapy suffered a major setback with the death of 18-year-old Jesse Gelsinger. Jesse was participating in a gene therapy trial for ornithine transcarboxylase deficiency (OTCD). He died from multiple organ failures 4 days after starting the treatment. His death is believed to have been triggered by a severe immune response to the adenovirus.
Another major blow came in January 2003, when the FDA placed a temporary halt on all gene therapy trials using retroviral vectors in blood stem cells. FDA took this action after it learned that a second child treated in a French gene therapy trial had developed a leukemia like condition. Both the child and another, who had developed a similar condition in August 2002, had been successfully treated by gene therapy for X-linked severe combined immunodeficiency disease (X-SCID), also known as “bubble baby syndrome”.
FDA’s Biological Response Modifiers Advisory Committee (BRMAC) met at the end of February 2003 to discuss possible measures that could allow a number of retroviral gene therapy trials for treatment of life-threatening diseases to proceed with appropriate safeguards. FDA has yet to make a decision based on the discussions and advice of the BRMAC meeting.
B. 4b. Problems with Gene Therapy:
Short-lived nature of gene therapy—before gene therapy can become a permanent cure for any condition, the therapeutic DNA introduced into target cells must remain functional and the cells containing the therapeutic DNA must be long-lived and stable. Problems with integrating therapeutic DNA into the genome and the rapidly dividing nature of many cells prevent gene therapy from achieving any long-term benefits. Patients will have to undergo multiple rounds of gene therapy.
Immune Response:
Anytime any foreign body is introduced into human tissues, the immune system is designated to attack the invader. The risk of stimulating the immune system in a way that reduces gene therapy effectiveness is always a potential risk. Further, the enhanced immune response to invaders makes it difficult for gene therapy to be repeated in patients.
Problems with Viral Vectors:
Viruses, the carrier of choice in most gene therapy studies, present a variety of potential problems to the patient such as toxicity, immune and inflammatory responses. In addition, there is always the fear that the viral vector, once inside the patient, may recover its ability to cause disease.
Multi-Gene Disorders:
Conditions or disorders that arise from mutations in a single gene are the best candidates for gene therapy. Unfortunately, some of the most commonly occurring disorders such as heart disease, high blood pressure, Alzheimer’s disease, arthritis, and diabetes are caused by the combined effects of variations in many genes. Multi-genic or multifactorial disorders such as these would be especially difficult to treat effectively using gene therapy.
C. Uncertainties with Animal Biotechnology:
As with new technologies, animal biotechnology faces a variety of uncertainties, safety issues and potential risks. For example, concerns have been raised regarding: the use of unnecessary genes in constructs used to generate transgenic animals, the use of vectors with the potential to be transferred or to otherwise contribute sequences to other organisms, the potential effects of genetically modified animals on the environment, the effects of biotechnology on the welfare of the animal, and potential human health and food safety concerns for meat or animal products derived from animal biotechnology. Before animal biotechnology will be used widely by animal agriculture production systems, additional research will be needed to determine if the benefits of animal biotechnology outweigh these potential risks.