Get the answer of: Does Z-DNA Play a Regulatory Role?
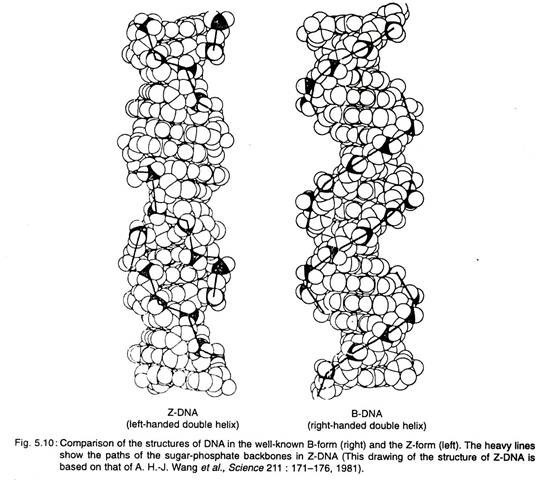
One of the more interesting discoveries of the last decade was that segments of DNA that have sequences in which purines and pyrimidines alternate along each strand can form left-handed double helices. The Watson-Crick B-form of DNA is a right-handed double helix. The novel left-handed double-helical form of DNA has been named Z-DNA for the zigzagged paths of the sugar-phosphate backbones of the molecule (Fig. 5.10).
Normally, the Z-form of alternating purine and pyrimidine DNA sequences occurs only at high salt concentrations. When some of the bases in the potential Z-form sequences are methylated however, the Z-conformation is stable at lower salt concentrations.
Thus, Z- DNA composed of alternating purine-pyrimidine sequences containing methylated bases may be stable in vivo. Moreover, its stability is enhanced by cations, including polyamines such as spermine, by negative supercoiling and by DNA binding proteins specific for Z- DNA.
In fact, there is evidence that Z-DNA exists in the interband regions of the giant salivary gland chromosomes of Drosophila melanogaster and in the transcriptionally active macronucleus of the ciliated protozoan Stylonychia mytilus. A. Rich and colleagues have prepared antibodies specific for Z-DNA, these antibodies do not react with B-form DNA.
Rich and coworkers have shown that the Z-DNA-specific antibodies bind to the interband regions of the polytene chromosomes of D. melanogaster. It will be of great interest to determine the sequences and methylation patterns of the DNA in the interband regions of these polytene chromosomes. Do these interband sequences control the expression (“puffing”) of the genes located in the adjacent bands? Or are these sequences merely structural or involved in synapsis?
Another hint of the possible involvement of Z-DNA in regulating gene expression is that the structures of certain regulatory proteins suggest that they may bind in the major groove of left-handed double helices, but not right-handed helices.
In fact, D. B. McKay and T. A. Steitz have proposed that the catabolite activator protein stabilizes its CAP-binding sequence in a left-handed conformation. They further propose that this right-handed to left- handed transition in the double helix unwinds the adjacent promoter or RNA polymerase binding site and thus activates transcription of the adjacent structural genes.
Repressor proteins might act in the opposite direction, stabilizing regulatory sequences in the right-handed B-form and preventing transcription. Although their functions are still unknown, Z- DNA-specific binding proteins have been isolated from Drosophila.
When alternating purine-pyrimidine sequences are complexed with histones, these sequences do not display B-DNA to Z-DNA transitions. This and other observations suggest that Z-DNA sequences must be stabilized by Z-DNA-specific binding proteins, and that Z-DNA sequences may not be found in nucleosomes.
Indications of nuclease sensitivity of B- DNA to Z-DNA junctions has led to speculation that such junctions may be related to the nuclease-sensitive sites near the promoter regions of transcriptionally active genes.
Clearly much more information is needed before the potential validity of these proposals can be evaluated. Although transitions from sequences of DNA in the B-conformation to sequences in the Z-conformation have been shown to occur within individual plasmids, the biological significance of these transitions remains unknown.
Experiments designed to test the possibilities:
1. That B-form to Z-form transitions in DNA are involved in the regulation of gene expression.
2. Regulatory proteins may act by binding to and stabilizing one or the other of these conformation may lead to some exciting developments during the next decades, hopefully.