In this article we will discuss about the morphology and multiplication of T-Even phages.
The dsDNA tailed phages of Caudovirales account for 95% of all the phages reported. Phages are classified by the International Committee on Taxonomy of Viruses (ICTV) according to morphology and nucleic acid as given in Table 18.1.
There are three even-numbered T-phages (e.g. T2, T4 and T6) and four odd-numbered T- phages (e.g. T1, T3, T5 and T7) that infect E.coil. Out of the seven coliphages of T-series, the virulent T4 phages is most extensively studied.
The T4 phage infects non-motile strain B of E. coli and forms plaques on growing colonies:
Morphology of T-Even Phages:
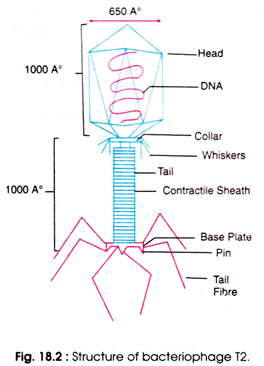
The T4 phage is tadpole shaped and consists of the five important sub-structures such as the head, head-tail connector, tail base plate and fibers (Fig. 18.2). The viral particle is naked icosahedral and tailed. The head is an elongated, bi-pyramidal, hexagonal like prism consisting of two 10-faced equatorial bands. It consists of a pyramidal vertex at either ends also. Size of head is 95 X 65 nm which consists of about 2,000 identical protein subunits (capsomers).
The phage consists of a long helical tail which is connected to the head with a connector having a collar with attached whiskers. Around the tail, fibres remain folded and held at midpoint by the whiskers. Size of the tail is 80 x 18 nm.
It consists of an inner tubular core (having a hole of 25 A) which is surrounded by a contractile sheath. Protein subunits (144) arranged in 24 rings each containing 6 subunits, constitute the sheath. The sheath connects the head at one end and base plate at the other end.
From the head at distal end, there is a hexagonal base plate attached to an end of tail. The base plate contains six spikes or tail fibers at its six corners. The spikes are 130 x 2 nm in size. The spikes have two parts, the proximal half fibre, and the distal half fibre. The former is attached to the base plate and the later helps in recognition of specific receptor sites present on cell surface of the bacterial cell wall.
A dsDNA molecule of about 50 µm is tightly packed inside the head. The DNA is about 1,000 times longer than the phage itself. It is circular and terminally redundant. Unlike other DNA, all the T-even phages contain 5-hydroxy-methyl-cytosin e instead of cytosine, so that synthesis of phage can occur easily.
Multiplication of T-Even Phages:
The multiplication processes of T-even phages are a little complex. It is accomplished in certain stages viz, adsorption, penetration, synthesis, replication, maturation (assembly) and release.
(i) Adsorption:
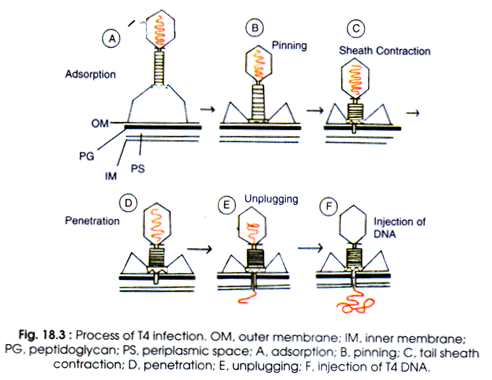
T4 phage particles interact with cell wall of E.coli. The phage tail makes contact between the two, and tail fibres recognise the specific recognition sites present on cell surfaces (Fig. 18.3A). However, any part of the bacterial cell may act as specific receptor sites e.g. flagella, pili, carbohydrates and proteins presents in the membrane or cell wall itself. Specific receptor in the bacterium is the lipopolysaccharide (LPS) or lipoprotein or both.
The LPS of tail fibres act as receptor in phages T3, T4 and T7, whereas lipoprotein acts as receptor in T2 and T6. In T5 phage both LPS and lipoprotein acts as receptors. When these receptors are absent, the bacteria become resistant to phage.
Therefore, it is not necessary that every interacting phage is adsorbed on bacterial cell. However, if phages are added artificially in excess, multiple adsorption of upto 200-300 phages per bacterium may occur.
When the contact is made between tail fibres and bacterium, it becomes unfolded from the tail. Unfolding and release of tail fibres from whiskers is governed by certain cofactors, for example tryptophan is needed (1 mg/ml) (by Benzer strain T4B), and in some T phages the absence of tryptophan (e.g. in Doerman strain T4D) and indole (e.g. in Hershey strain T2H) are not required. High concentration (0.2-0.6 M) of glucose is inhibitory for adsorption of T4 to bacterial cell.
The whole process of interaction of phage to recognition of specific receptor site is known as landing (Fig. 18.3A). After landing there starts the second process of adsorption which is known as pinning (B). Before pinning the phage can move attached with tips of tail fibres to cell surface until it finds the site for pinning of spikes.
Pinning is the irreversible process. All the activities before pinning are reversible. Possibly it occurs at the point where both plasma membrane and cell wall are attached.
(ii) Penetration:
The process of penetration occurs by mechanical and also enzymatic digestion of cell wall. At the recognition site phage either digests certain cell wall structure by viral enzymes e.g. lysozyme or activates the degradative enzymes of the host. But the real penetration of phage into the host is mechanical.
After pinning the tail sheath contracts and, therefore, appears shorter and thicker (Fig.18.3C, Fig.18.4A). Sheath contracts by rearrangement of discs from 24 to 12. The phage uses its energy in tail contraction as ATP, and calcium has been detected in purified T-even phages. The activity of phage contraction is comparable to that of micro-syringe.
Kollenberger (1970) has described very nicely the process of contraction of hollow stylus into the bacterial cell through the central hole in the base plate (D). The base plate through the centre enlarges after contraction of sheath. The stylus can penetrate the cell wall but not the plasma membrane.
After making contact with a component of plasma membrane (possibly phosphatidyl glycerol) unplugging of phage DNA begins (E). Thereafter, DNA is injected into the cell without requiring metabolic energy (F).
Phage head and tail remain outside the cell. Such empty protein coats are known as ‘ghosts’. Phage T1 and T5 do not contain contractile sheath, even then penetration of tail and injection of DNA into the bacterial cell occur.
(iii) Synthesis:
The process of synthesis of macromolecules leading to phage multiplication involves the degradation of bacterial chromosomes, protein synthesis, and DNA replication.
(a) Degradation of bacterial chromosome:
After infection by T- even phage, the bacterial chromosome is unfolded due to relax of super-helical twisting. The nucleoid, which is present in the centre, is disrupted and DNA helix is attached with plasma membrane at different points. This process is facilitated by nucleoid disrupting gene (T4 dna gene) products.
The RNA polymerase-I binds to membrane simultaneously (Fig. 18.4 C). After 3 minutes of infection, protein synthesis by bacterial host stops, and partial break down of bacterial chromosome occurs. However, before 3 minutes of infection replicative synthesis of bacterial cell takes place.
However, upto 10 minutes non-conservative DNA synthesis occurs just to repair the damage in the bacterial DNA caused by nucleases -encoded by phage. The RNA polymerase and ribosomes are modified which initiate protein synthesis of phage but not of bacterial cell.
(b) Protein synthesis:
The phage DNA contains early and late genes, therefore, after a few minutes of introduction of genome bacterial mRNA and protein synthesis stop. The early and late genes transcribe (at different intervals) the early mRNA and late mRNA respectively. After 5 minutes of infection the early mRNA of phage have been detected in bacterial cell.
Late mRNA synthesis starts after 10 minutes of infection. After 20 minutes both the early and late mRNAs are found in the host cell. The left strand of T4 DNA transcribes the early genes and the right strand of DNA transcribes the late genes. The early genes are of two types, immediate early and delayed early genes.
The immediate early genes are transcribed after 1.25 or 2.5 minutes of infection by using the bacterial RNA polymerase. These genes also code enzymes that degrade bacterial chromosome. The delayed early genes transcribe the mRNA from 2.5 to 3.75 minutes.
The delayed early genes synthesize the phage enzymes which produce 5-hydroxy-methyl-cytosine (5HMC) by using cytosine of the bacterial DNA. At this stage the bacterial restriction enzymes could not degrade the phage genome.
Use of chloramphenicol can warrant the transcription of the delayed early mRNA. The late phage gene products are the lysozyme and the structural components of new phage particles such as the heads, tails and fibres.
(c) Replication of T4 DNA:
During the first 10 minutes after infection of the phage DNA, no phage could be recovered from the infected bacterium. This time interval is called as eclipse period (Fig. 18.5). During the eclipse phase, no infectious phage particles can be found either inside or outside the bacterial cell.
The phage nucleic acid takes over the host biosynthetic machinery and phage specified RNAs and proteins are synthesized. Early mRNAs code for early proteins which are needed for phage DNA synthesis and for shutting off host DNA, RNA and protein synthesis. After synthesis of phage DNA late mRNAs and late proteins are synthesized.
The late proteins are the structural proteins that comprise the phage as well as the proteins needed for lysis of the bacterial cell. Moreover, replication of T4 DNA is a complex process. T4 DNA differs from the typical DNA molecules as the former contains no cytosine, but consists of modified base and 5-HMC which pairs with guanine. Several glucose like sugars are covalently linked to the 5-HMC.
There are five important events of T4 DNA replication:
(a) Degradation of host DNA,
(b) Synthesis of 5-HMC,
(c) Prevention of incorporation of cytosine into the T4 DNA,
(d) Glucosylation of T4 DNA, and
(e) Enzymology of DNA replication.
Generally, the bacterial DNA is degraded by phage coded exonuclease to deoxyribonucleotide monophosphate (dNMP). Then the dNMP built up dATP, dTTP, dGTP and dCTP by the bacterial enzymes which provide sufficient dNTP to synthesize 30 T4 DNA molecules.
Therefore, replacement of cytosine to 5-HMC is done which becomes glucosylated. This modified nucleotide is protected from degradation by the bacterial restriction enzyme. Timing of life cycle in minutes at 37°C is outlined in Table 18.3. 

After 6 minutes of infection, replication of T4 DNA begins, then the rate of replication reaches maximum after 10-12 minutes. DNA synthesis is not done by T4 DNA polymerase, but a short RNA primer is synthesized which presumably starts each step of DNA synthesis. On RNA primer T4 DNA polymerase extends the DNA strand by using one of the strands as template. DNA polymerase possesses 3′ exonuclease activity but lacks 5′ exonuclease activity.
Gene 32 encodes the T4 DNA binding proteins which is required for replication and recombination. It maintains the DNA in single stranded state. The genetic map of T4 phage is given by King (1968) (Fig.18.6).
It has been found that the different regions in phage genome regulate the specific operations like promotion of synthesis of viral DNA and of RNA molecules (King, 1968). T4 DNA replicates by rolling circle mechanism also. After viral DNA synthesis, the late proteins are formed.
(iv) Assembly (Maturation):
When the structural proteins and phage DNA are synthesized, assembly of these components begins to give rise the mature phages. About 135 T4 DNA molecules are identified which account for 90% of the DNA. About 20% genes are to be identified and 50 genes take part in morphogenesis of this phage.
The process of assembling the phage particles is known as maturation. Assembly of phage components begins in a well defined order, such as the head, tail, and tail-fibres synthesized independently, are combined to yield phage particles (Fig. 18.7). After 20 minutes of infection about 300 new phages have been assembled. Some steps in assembly of coliphage T4 have been described by Edger and Lielausis (1968).
J.D. Watson and F.C. Crick (1972) postulated that the assembly of preformed parts into a virion is a process of condensation, and the shape and design of a viral capsid is determined by the specific bonding properties of its identical capsomers. However, it appears that the maturation is a sequential event in which each step requires the previous ones.
(v) Lysis and Release:
After eclipse period maturation of phage particles starts where they accumulate inside the bacterial cell. The progenies are released by the sudden bursting i.e. lysis of host cell wall. Lysozyme is attached to the tip of the tail of phages.
Time taken from infection to lysis is known as latent period. Yield of phage per bacterium is known as burst size. Starting with infection by a phage of the bacterial cell to release the newly synthesized viral particles with respect to the time is known as one step growth curve (Fig 18.5). This growth curve was first introduced by Ellis and Delbrack (1939).