Let us make an in-depth study of the protein synthesis. After reading this article you will learn about: 1. Protein Synthesis 2. Components of Protein Synthesis 3. Mechanisms of Protein Synthesis and 4. Initiation of Protein Synthesis.
Contents
Protein Synthesis:
Proteins are giant molecules formed by polypeptide chains of hundreds to thousands of amino acids. These polypeptide chains are formed by about twenty kinds of amino acids. An amino acid consists of a basic amino group (-NH2) and an acidic carboxyl group (-COOH). Different arrangement of amino acids in a polypeptide chain makes each protein unique.
Proteins are fundamental constituents of protoplasm and building material of the cell.
They take part in the structural and functional organization of the cell. Functional proteins like enzymes and hormones control the metabolism, biosynthesis, energy production, growth regulation, sensory and reproductive functions of the cell. Enzymes are catalysts in most of the biochemical reactions. Even the gene expression is controlled by enzymes. The replication of DNA and transcription of RNA is controlled by the proteinous enzymes.
Components of Protein Synthesis:
Protein synthesis is governed by the genetic information carried in the genes on DNA of the chromosomes.
The main components of the protein synthesis are:
1. DNA
2. Three types of RNAs
3. Amino acids
4. Ribosomes
5. Enzymes.
DNA is the master molecule which posseses the genetic information about the sequence of amino acids in a polypeptide chain. Structure and properties of DNA regulate and control the synthesis of proteins.
DNA present in the nucleus sends out information in the form of messenger RNA into the cytoplasm, which is the site of the protein synthesis in eukaryotes. The messenger RNA carries the information regarding the sequence of amino acids of the polypeptide chain to be synthesized. This message or information is in the form of a genetic code. This genetic code specifies the language of amino acids to be assembled in a polypeptide.
The genetic code is deciphered or translated into a sequence of amino acids.
Composition of Genetic Code:
DNA molecule has three components. They are sugar, phosphates and nitrogen bases. Only nitrogen base sequence varies in different DNA molecules. Thus, the sequence of nitrogen bases or nucleotides in a DNA segment is the code or language in which the DNA sends out the message in the form of messenger RNA (mRNA).
The mRNA carries the genetic message (genetic code) in the form of nucleotide sequence. It has been found that there is colinearity between nucleotide sequence of mRNA and amino acid sequence of the polypeptide chain synthesized.
The genetic code is the language of nitrogen bases. There are four kinds of nitrogen bases and twenty kinds of amino acids. Therefore four-letter language of nitrogen bases specifies the twenty letter language of amino acids.
Mechanisms of Protein Synthesis:
In prokaryotes, the RNA synthesis (transcription) and protein synthesis (translation) take place in the same compartment as there is no separate nucleus. But in eukarytoes, the RNA synthesis takes place in the nucleus while the protein synthesis takes place in the cytoplasm. The mRNA synthesized in the nucleus is exported to cytoplasm through nucleopores.
First, Francis Crick in 1955 suggested and later Zemecnik proved that prior to their incorporation into polypeptides, the amino acids attach to a special adaptor molecule called tRNA. This tRNA has a three nucleotide long anticodon which recognizes three nucleotide long codon on mRNA.
Role of Ribosomes in Protein synthesis:
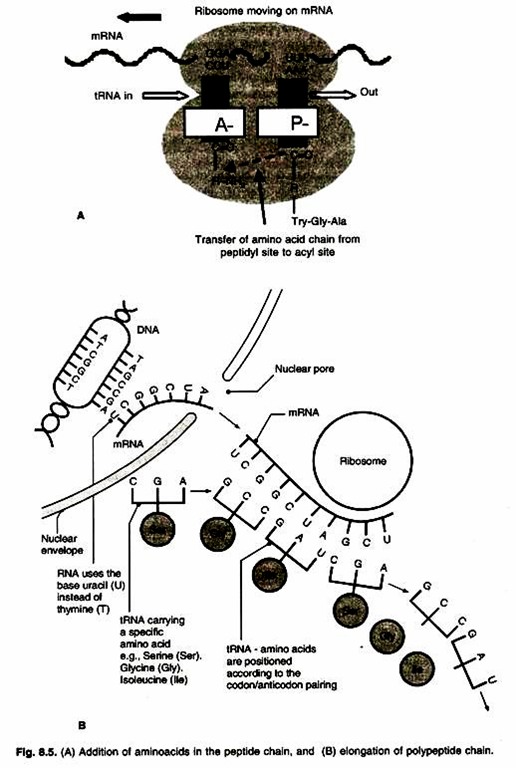
Ribosome is a macromolecular structure that directs the synthesis of proteins. A ribosome is a multicomponent, compact, ribonucleoprotein particle which contains rRNA, many proteins and enzymes needed for protein synthesis. Ribosome brings together a single mRNA molecule and tRNAs charged with amino acids in a proper orientation so that the base sequence of mRNA molecule is translated into amino acid 1 sequence of polypeptides.
Ribosome is a nucleoprotein particle having two subunits. These two subunits lie separately but come together for the synthesis of polypeptide chain. In E. coli ribosome is a 70S particle having two subunits of 30S and 50S. Their association and dissociation depends a upon the concentration of magnesium.
Small subunit of ribosome contains the decoding centre in which charged tRNAs decode o the codons of mRNA. Large subunit contains peptidyl transferase centre, which forms the peptide bonds between successive amino acids of the newly synthesized peptide chain.
Both 30S and 50S subunits consist of ribosomal RNA (rRNA) and proteins.
The mRNA binds to the 16S rRNA of smaller subunit. Near its 5′-end mRNA binds to the 3′-end of 16S rRNA.
The main role of ribosome is the formation of peptide bond between successive amino acids of the newly synthesized polypeptide chain. The ribosome has two channels in it. The linear mRNA enters and escapes through one channel, which has the decoding centre. This channel is accessible to the charged tRNAs. The newly synthesized polypeptide chain escapes through the other channel.
Direction of Translation:
Each protein molecule has an -NH2 end and -COOH end. Synthesis begins at amino end and ends at carboxyl end. The mRNA is translated in 5 → 3′ direction from amino to carboxyl end. Synthesis of mRNA from DNA transcription also occurs in 5′ → 3′ direction.
Initiation of Protein Synthesis:
Formation of Initiation Complex:
First of all 30S subunit of the 70S ribosome starts initiation process. The 30S subunit, mRNA and charged tRNA combine to form pre-initation complex. Formation of pre-initiation complex involves three initiation factors IF1, IF2 and IF3 along with GTP (guanosine triphosphate). Later 50S subunit of ribosome joins 30S subunit to form 70S initiation complex.
Information for protein synthesis is present in the form of three nucleotide codons on mRNA. Protein coding regions on mRNA consist of continuous, non-overlapping triplet codons. The protein coding region on mRNA is called open reading frame which has a start codon 5′-AUG-3′ and a stop codon in the end. Each open reading frame specifies a single protein. Prokaryote mRNA has many open reading frames, therefore encode multiple polypeptides and are called polycistronic mRNAs.
Near the 5′-end of mRNA lies the start codon which is mostly 5′-AUG-3′ (rarely GUG) in both prokaryotes and eukaryotes. Ribosome binding site (RBS) in prokaryotes lies near the 5′- end of mRNA ahead (upstream) of AUG codon.
Between 5′-end and AUG codon there is a sequence of 20-30 bases. Of these, there is a sequence 5′-AGGAGGU-3′. This purine rich sequence is called Shine-Dalgarno sequence and lies 4-7 bases ahead (upstream) of AUG codon.
The 3′-end region of 16S rRNA is 30S subunit has a complementary sequence 3′-AUUCCUCCA-5′. This sequence forms base pairs with Shine-Dalgarno sequence for binding of mRNA to ribosome. Shine-Dalgarno sequence is the ribosome binding site (RBS). It positions the ribosome correctly with respect to the start codon.
There are two tRNA binding sites on ribosome covering 30S and 50S subunits. The first site is called “P” site or peptidyl site. The second site is called “A” site or aminoacyl site. Only the initiator tRNA enters the “P” site. All other tRNAs enter the “A” site.
For every amino acid, there is a separate tRNA. The identity of a tRNA is indicated by superscript, such as tRNAArg (specific for amino acid Arginine). When this tRNA is charged with amino acid Arginine, it is written as Arginine-tRNAArg or Arg-tRNAArg. Charged tRAN is called aminoacylated tRNA.
In bacteria, the first amino acid starting the protein is always formyl methionine (fMet). When AUG appears as the start codon on mRNA only fMet is incorporated. The tRNA molecule carrying formyl methionine is called tRNA™61. Therefore the first initiator charged aminoacyl tRNA is always fMet-tRNAfMet. When AUG codon is encountered in the internal location (other than the start codon), methionine is not formylated and tRNA carrying this methinine is tRNAmMet.
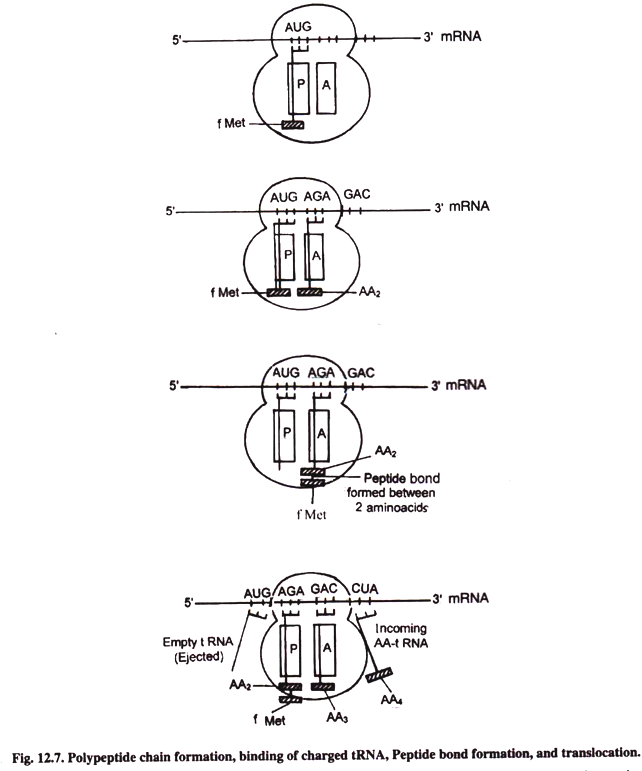
First of all the charged initiator tRNA called tMet-tRNAfMet occupies the “P” site on ribosome. This position brings its anticodon and start codon AUG of mRNA together in such a way that the anticodon of charged tRNA and codon of mRNA form base pair with each other. Thus reading or translation of mRNA begins.
The “A” site is available to the second incoming charged tRNA whose anticodon forms base pairs with the second codon on mRNA.
Charging of tRNA:
Attachment of amino acids to tRNAs is called charging of tRNA. All tRNAs at their 3′-terminus have a sequence 5′-CCA-3′. At this site amino acids bind with the help of enzyme aminoacyl tRNA synthatase. Charging of tRNA occurs in two steps.
1. Activation of amino acids:
Energy molecule ATP activates the amino acids. This step is catalysed by specific activating enzymes called aminoacyl tRNA synthatases. Every amino acid has a separate enzyme AA-RNA synthatase enzyme.
2. Transfer of amino acids to tRNA:
AA-AMP enzyme complex reacts with a specific tRNA and transfers the amino acid to tRNA, as a result of which AMP and enzyme are set free.
This first AA-tRNA is fMet-tRNAfmet which is amino acid formyl methionine bound to tRNA. This fixes itself to “P” site on ribosome. After this the second AA-tRNA attaches itself to “A” site on ribosome. In this way polypeptide chain elongation begins.
Polypeptide Chain Elongation:
Polypeptide chain elongation requires some elongation factors. These elongation factors are Tu and G.
EF-Tu forms a complex with AA2-tRNA and GTP and brings it to the “A” site of ribosome. Once the AA2-tRNA is in place at “A” site, the GTP is hydrolysed to GDP and EF- Tu is released from the ribosome. EF-Tu-GTP complex is regenerated with the help of another factor Ts.
Formation of Peptide Bond:
The main role of ribosome is to catalyse the formation of peptide bonds between successive amino acids. In this way amino acids are incorporated into protein.
Now both “P” site and “A” site on ribosome are occupied by charged tRNAs having amino acids. Peptide bond is formed between two successive amino acids at “A” site. It involves cleavage of bond between f-Met and tRNA. This is catalysed by the enzyme tRNA deacylase.
Peptide bond is formed between the free carboxyl group (-COOH) of the first amino acid and the free amino group (- NH2) of the second amino acid at the “A” site. The enzyme involved in this reaction is peptidyl transferase. After the formation of peptide bond, between two amino acids, the tRNA at “P” site becomes uncharged or deacylated and tRNA at “A” site now carries a – ill protein chain having two amino acids. This occurs in 50S subunit of ribosome.
The peptidyltransferase which catalizes the peptide bond formation between successive amino acids consists of several proteins and molecule of 23S rRNA in the ribosome. This 23S rRNA is a ribozyme.
Translocation:
The peptidyl tRNA carrying two amino acids present at “A” site is now translocated to”P” site. This movement is called translocation. Elongation factor called EF-G control translocation. This factor G is called translocase. Hydrolysis of GTP provides energy for translocation and release of deacylated tRNA (free of amino acid).
Translocation also involves movement of ribosome along mRNA towards its 3′-end by a distance of one codon from first to second codon. This movement shifts the dipeptidyl tRNA (carrying two amino acids) from “A” to “P” site.
In addition to these two sites P and A, a third site “E” (exit site) on 50 S ribosome is present. Deacylated tRNA (deprived of amino acid) moves for “P” site to “E” site from where it is ejected out.
Then the third amino acid (next amino acid) charged on tRNA comes to lie in now empty site “A”. Then dipeptidyl chain having two amino acids present on P site form peptide bond with the third amino acid at “A” site. Then the three amino acid chain is translocated to “P” site. Now the polypeptide chain has three amino acids. This elongation process goes on and on. At each step a new amino acid is added to the polypeptide chain. After each elongation, ribosome moves by one codon in 5′ → 3′ direction.
Chain Termination:
The presence of termination codons or stop codons on mRNA causes the polypeptide chain to be terminated. Synthesis stops when elongation chain comes across stop codons on “A” site. The stop codons are UAA, UGA and UAG. There is no tRNA which can bind these codons.
There are three release factors in prokaryotes, which help in chain termination. They are RF1, RF2 and RF3.
Polyribosome or Polysome:
A single mRNA molecule can be read simultaneously by several ribosomes. A polyribosome or polysome consists of several ribosomes attached to the same RNA. The number of ribosomes in a polysome depends upon the length of mRNA.
A fully active mRNA has one ribosome after every 80 nucleotides. There may be about 50 ribosomes in a polycistronic mRNA of prokaryotes. Ribosomes move along mRNA in 5′ 3′ direction. There is a gradual increase in the size of polypeptide chain as the ribosomes move along mRNA towards its 3′-end. Polypeptide chain starts near the 5′-end and is completed near the 3′-end.
The ribosomes closest to the 5′-end of mRNA have the smallest polypeptide chain, while ribosomes nearest to the 3′-end have longest chain. Polysome increases the rate of protein synthesis tremendously. In bacteria protein is synthesized at the rate of about 20 amino acids per second.
Simultaneous Transcription and Translation in Prokaryotes:
In prokaryotes, all components of transcription and translation are present in the same compartment. The mRNA molecule is synthesized in 5′ → 3′ direction and protein synthesis also occurs in 5′ → 3′ direction. In this way mRNA molecule while still under synthesis has a free 5′-end whose other end is still under synthesis.
Ribosomes bind at free 5′-end and start protein synthesis. In this way the free end (5′-end) of mRNA starts the process of protein synthesis while still attached to DNA. This is called Coupled Transcription and Translation. This increases the speed of protein synthesis. After the protein synthesis is completed, the degradation of mRNA molecule by nucleases also starts at 5′-end and proceeds in 5′ → 3′ direction.
Protein Synthesis in Eukaryotes:
Protein synthesis in eukaryotes is basically similar to that of prokaryotes except some differences.
The ribosomes in eukaryotes are of 80S having 40S and 60S subunits. In eukaryotes the initiating amino acid is methionine and not f-methionine as in the case of prokaryotes. A special tRNA binds methionine to start codon AUG. This tRNA is called tRNAiMet. This is distinct from tRNAMet which binds amino acid methionine to any other internal position in the polypeptide.
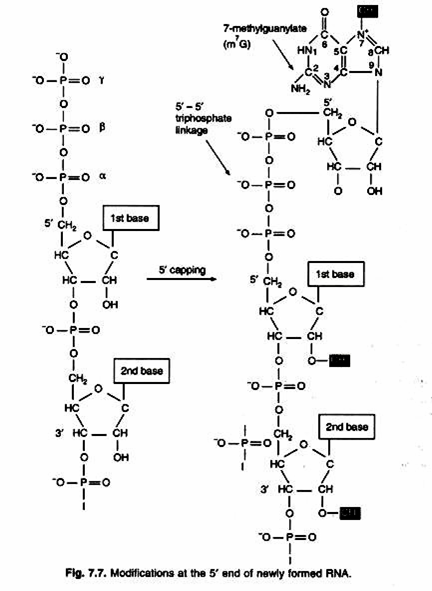
There is no Shine-Dalgarno sequence in eukaryotic mRNA to function as ribosome binding site. Between 5′-end and AUG codon of mRNA there is a sequence of bases called cap. Small subunit of ribosome scans the mRNA in 5′ → 3′ direction until it comes across 5′- AUG-3′ codon. This process is called scanning. Initiation factors also closely associated with 3′-end of mRNA through its poly-A tail. Initiation factors circularize mRNA by its poly-A tail. In this way poly-A tail also contributes to the translation of mRNA. Eukaryotic mRNAs are monocistsonic and encode a single polypeptide, therefore have a single open reading frame.
There are ten initiation factors in eukaryotes. They are elF (eukaryotic intiation factors) are elFI, eIF2, eIF3, eIF4A, eIF4B, eIF4C, eIF4D, eIF4F, eIF5, eIF6.
There are two elongation factors in eukaryotes like prokaryotes. They are eEFl (similar to EF-Tu) and eEF2 (similar to EF-G).
Eukaryotes have only one release factor eRF which requires GTP termination of protein synthesis. It recognizes all the three stop codons.
In eukaryotes the mRNA is synthesized in the nucleus, then processed, modified and passed on into the cytoplasm through nucleopores. The protein synthesis takes place in the cytoplasm. The mRNA in prokaryotes is very unstable and its life span is of a few minutes only. The mRNA of eukaryotes is quite stable and has a longer life span extending upto several days.
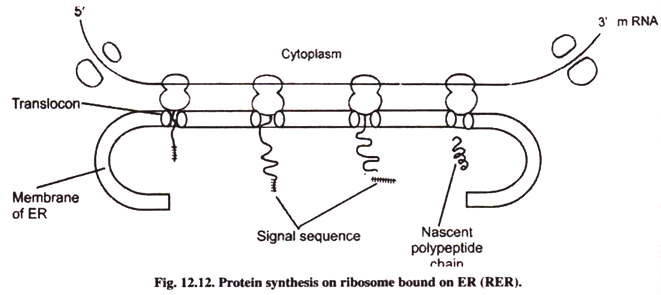
Protein Synthesis on Bound Ribosomes:
Ribosomes occur in free state in the cytoplasm as well as bound to the outer surface of endoplasmic reticulum called rough endoplasmic reticulum (RER). The attachment of ribosomes to ER occurs after the protein synthesis starts. Whether the ribosomes synthesize protein on free or attached state depends upon the type of proteins to be synthesized by ribosomes. Most of the proteins which remain in free state in the cytoplasm are synthesized by free ribosomes.
Proteins synthesized by ribosomes on ER enter into the lumen of cisternae of ER from where they may enter into golgi apparatus where they are glycosylated and form secretary granules and many of them enter lysosomes.
Modification of Folding of Released Polypeptides:
DNA molecule specifies only the primary structure while folding and other modifications controlled by proteins themselves.
The newly synthesized polypeptide is not always a functional protein. The newly released polypeptide may undergo various modifications. An enzyme deformylase removes the formyl group of first amino acid methionine. The cleavages of proteins are most common. Some enzymes like exo-amino-peptidases remove some amino acids either from N-terminus end or from C-terminus end or both ends.
Internal amino acids may also be removed as in the case of insulin. Polyproteins are cleaved to generate individual proteins. The polypeptide chain singly or in association with other chains may fold up to form tertiary or quaternary structures. Prosthetic groups join many proteins. Some proteins assist in folding up of polypeptides. They are called chaperone proteins or chapronin proteins. Examples are Bacterial gro EL (E. coli), mitochondrial hsp 60 mitonin.
Various chemical common modifications of newly released proteins are glycosylation, phosphorylation, methylation, acetylation etc.
Protein Sorting or Protein Trafficking or Protein Targeting:
The proteins synthesized in the cell have to be translocated to the nucleus or other target organelles. Newly synthesized polypeptides have a signal sequence (which is a polypeptide) consisting of 13-36 amino acids. It is known as leader sequence. This signal sequence is recognized by receptors located within the membranes of the target organelles.
When proteins are synthesized on free ribosomes, the transfer takes place after the translation. When the protein synthesis takes place on ribosomes attached to the endoplasmic reticulum (Rough ER), the transfer takes place simultaneously with translation and is called co-translational transfer. The proteins which enter into the lumen of rough ER may enter into golgi apparatus, from where they may enter secretary lysosomes. The signal sequence is degraded by protease enzymes.
Once all these proteins are assembled into their proper place, they provide the proper biochemical machinery, which keeps the cell feeding, locomoting, multiplying and alive.
Inhibitors of Protein Synthesis:
There are many chemicals, both synthetic as well as those obtained from different sources like fungi, which bind to the components of translation machinery and arrest the translation process. Most of them are antibacterial agents or antibiotics that act exclusively on bacteria and are thus powerful tools in the hands of man to combat various infectious diseases. Most of antibiotics are inhibitors of translation machinery.
Puramycin:
It binds at “A” site on ribosome. This causes pre-mature termination of polypeptide chain.
Kirromycin:
It inhibits the elongation factor EF-Tu.
Fusidic acid:
It inhibits the elongation factor EF-G.
Tetracycline:
It attacks “A” site on ribosome and prevents the binding of aminoacyl- tRNA.
Chloramphenicol: It blocks the peptidyl transfer reaction.
Erythromycin:
It binds the polypeptide exit channel of ribosome, therefore blocks the exit of growing polypeptide chain, thus stops the translation process.
Streptomycin and Neomycin:
These inhibit the binding of tRNAfMet to the “P” site.
Inhibitors in Eukaryotes:
Diphtheria toxin is a toxin produced by corynebacterium diphtheriae. This causes modification of eukarotic elongation factor.