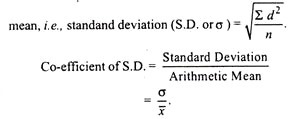
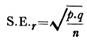Let us make an in-depth study of the molecular self-assembly of nanotechnology. The below given article will help you to learn about the following things:- 1. Introduction to Nanotechnology Self-Assembly 2. The Five Factors Responsible for Self-Assembly 3. Examples of Controlled Self-Assemblies and 4. Conclusions and Discussion.
Contents
Introduction to Nanotechnology Self-Assembly:
Self-assembly is a process in which a set of components or constituents spontaneously forms an ordered aggregate through their global energy minimization. A vast number of macromolecules (like proteins, nucleic acid sequences, micelles, liposomes, and colloids) in nature adapt their final folding and conformation by self-assembly processes. There exist many examples in the literature of natural self-assemblies which occur spontaneously due to the forces of nature.
Examples of such natural self-assemblies are observed at all levels, from the molecular to macromolecular scale and in various living systems. Protein folding, diamondoids as host-guest chemistry molecular receptors, DNA double helix, formation of lipid bilayers, formation of micelles, micelle coacervates and steric colloids are some examples in which self-assembly is the dominant phenomenon.
Nanotechnology self-assembly encompasses a wide range of concepts and structural complexities, from the growth of crystals to the reproduction of complete biological entities. It is the aim of nanotechnology to master and design well controlled self-assemblies starting from various MBBs (Molecular building blocks). By utilizing the natures help in such self-assemblies one could form and produce various Nano structures and then larger systems and materials with the desired physicochemical properties.
Such larger heterogeneous aggregates should be able to perform intricate functions, or constitute new material forms, with unprecedented properties. This could lead to the creation of objects able to find their place on particular substrates or within given infrastructures. Directed self-assembly to design the desired synthetic nanostructures from MBBs is the main goal of nanotechnology.
Undoubtedly to achieve this goal it is necessary to utilize the knowledge about intermolecular interactions between MBBs, nanostructures steric arrangements, computer based molecular simulations, and biomimetic. Biomimetic is defined as making artificial products that mimic the natural ones based on the study of structure and function of biological substances.
The Five Factors Responsible for Self-Assembly:
There are five factors responsible for the success of a self-assembly process.
They include:
1. Molecular building blocks;
2. Intermolecular interactions;
3. Reversibility (or adjustability);
4. Molecular mobility (due to mixing and diffusion);
5. Process medium (or environment).
1. The Role of Molecular Building Blocks (MBBs) in Self-Assembly:
The self-assemblies of major interest in nanotechnology are mostly among larger size molecules, known as the molecular building blocks (MBBs), in the size ranges of 1-100 ф[nm]. This is because larger well-defined MBBs offer greater level of control over other molecules, and over the interactions among them that makes fundamental investigations especially tractable.
2. The Role of Intermolecular Interactions in Self-Assembly:
Forces responsible for self-assembly are, generally, weak non- covalent intermolecular forces including hydrogen bonding, electrostatic, van der Waals’, polar hydrophobic and hydrophilic interactions. To assure consistency and stability of the whole self-assembly complex the number of such weak interactions for self-assembly formation in each region of molecular conformation must be rather high. An example of a stable self-assembly made up of weak interactions is the second helical beta-sheet structure of proteins formed through hydrogen bonding as shown in Fig. 8.1.
3. Reversibility:
The on-going and other envisioned self-assembly processes in nanotechnology are controlled, but spontaneous, processes during which the MBBs associate and produce the desired ordered assemblies or aggregates. To achieve spontaneity it is necessary to run the process reversibly.
4. Molecular Mobility:
Due to its dynamic nature, a self-assembly process needs a fluid medium to proceed. Liquids, gases, supercritical fluids and the fluid side of the interfaces between a solid and a fluid phase are the possible environments for self-assembly. In all such cases, to achieve self-assembly, dynamic exchanges toward reaching the minimal energy level has to take place.
5. Process Medium:
A self-assembly is largely influenced by the environment in which it occurs. A molecular aggregate which is formed by the self-assembly process, is generally an ordered array which is thermodynamically the most stable conformation. Self-assembly occurs in liquid medium, in gaseous medium (including dense gases-supercritical fluid medium), near the interface between a solid and a fluid, or at the interface between a gas and a liquid.
Examples of Controlled Self-Assemblies:
The principles behind achieving successful controlled self-assemblies in the laboratory are presented above. Now it is appropriate to introduce a number of successfully achieved examples of self-assemblies.
There are two kinds of self-assembles based on the process medium:
A. Those which occur on the interface of a fluid and a solid phase.
B. Those which occur in the bulk fluid phase. The fluid phase can be a liquid, a vapor or a dense (supercritical) gas.
A. Self-Assembly Using Solid Surfaces-Immobilization Techniques:
There are a number of self-assembly methods which are achieved in the laboratory employing fluid medium as their association environment and a solid surface as their assembling nucleation and growth. An important first step in such self-assemblies is the immobilization of the assembly seed on solid surfaces. There is a number of immobilization techniques used for this purpose, some of which are reported here.
Immobilization of molecules as assembly seed on solid supports, which need to be used for self-assembly, can be achieved via covalent or non-covalent bonds between the molecule and the surface. Covalent bonds produce irreversible, thus stable, immobilization at all stages. Immobilization through non-covalent bonds is a reversible process and unstable at the onset of the process. However, it achieves stability upon appreciable growth of the assembly process.
A more common covalent bond used for immobilization is a sulfide bond and a noble metal. One such example is the covalent bond between thiol-bearing molecules (life alkane- thiol chains or proteins with cystine in their structure) and gold.
Some common non- covalent bonds used for immobilization are the following three kinds of bond:
(i) Affinity coupling via antibodies,
(ii) Affinity coupling by biotin-streptavidin (Bio-STV) system and its modification,
(iii) Immobilized metal ion complexation (IMIC).
These three kinds of non-covalent bonds are explained in more detail.
A-1. Affinity coupling via antibodies:
Antibody is a protein produced by the body’s immune system in response to a foreign substance (antigen). Antibodies are glycoproteins that are called immune-globulins that are found in the blood and tissue fluids produced by cells of the immune system. For direct immobilization, the purified antibodies are attached to gold substrates by using a bi-functional reagent with the thiol group on one side (Fig. 8.2). Highly oriented antibody immobilization can be achieved by assembling protein on gold surfaces via an introduced cystein-residue.
A-2. Affinity coupling by Biotin-Streptavidin (Bio-STV) System and its modification:
Avidin is a basic glycoprotein of known carbohydrate and amino acid content. Avidin combines stoichometrically with biotin (a vitamin B), which is a small molecule with high affinity to avidin. Streptavidin (STV) is a tetrameric protein, and like avidin, has four high affinity binding sites for biotin.
STV is a protein homologous to the avidin of yolk, produced by a strain of bacteria. STV binds very tightly to biotin, producing biotin-streptavidin (Bio-STV) complex. Furthermore streptavidin is not a glycoprotein and therefore does not bind to lectins as does avidin. The physical properties of streptavidin therefore make this protein much more desirable for use in immobilization systems than avidin. Because of its high affinity constant between the interacting partners, the biotin-avidin system is one of the most prominent systems used for immobilization.
Affinity coupling by Bio-STV system is one of the most efficient approaches to construct semi-synthetic nucleic acid-protein conjugates. Bio-STV affinity constant is about 1014 dm3moH, which is the strongest known ligand-receptor interaction. This method provides the possibility for immobilization of many molecules.
A–3. Immobilized Metal Ion Complexation (IMIC):
The IMIC method is based on the non-covalent binding of a biomolecule by complex formation with metal ions. Such metal ions are immobilized by chelators (molecules that bind to metal ions), like iminodiacetic acid [HOOC-CH2 NH-CH – COOH] or nitrilotriacetic acid [HOOC— CH2-N-(CH2-COOH)2] also called NTA (Figure 8.3).
In contrast to other affinity-based immobilization techniques, the interaction is based on low-molecular weight compounds. First, the chelator is attached to a surface, followed by loading with divalent metal cations (e.g. Ni2+, Zn2+, Cu2+). The tetra dental ligand NTA forms a hexagonal complex with the central Me2+, occupying four of the six coordination sites. Two sites are then available for binding biomolecules with electron donation groups coordinately.
Crystalline bacterial cell surface layers can be exploited for immobilization using appropriate chemical and physical processes. DNA oligomers can be used for site-selective immobilization of macromolecules. This is quite noteworthy, because it becomes feasible to direct self-assembly process on a solid surface via DNA directed immobilization (DDI).
Firstly, a DNA strand tags the desired site of the molecule and then the complementary DNA strand is fixed on a solid surface. Thus, completely specific DNA hybridization is exploited to immobilize macromolecules with controlled steric orientation. This method has been successfully performed to immobilize gold nanoparticles.
Fig. 8.4 depicts the role of DNA microarrays for site-selective immobilization of oligonucleotide-modified gold nanoparticles. To achieve this, a capture oligonucleotides’, cA, is first attached to a solid glass support to produce DNA microarrays. On the other side, the gold nanoparticles are tagged by several oligonucleotides, A, which are complementary to the capture oligonucleotide cA. Such DDI techniques allow for highly efficient, reversible and site-selective functionalization of laterally micro-structured solid supports.
These methods are particularly suitable for fabrication of reusable biochips and other miniaturized sensor devices containing biological macromolecules, enzymes and immunoglobulin’s.
A-4. Self-assembled monolayer (SAM):
A self-assembled monolayer (SAM) is defined as a two-dimensional film with the thickness of one molecule that is attached to a solid surface through covalent bonds. There are many applications in nanotechnology for SAM including nanolithography, modification of adhesion and wetting properties of surfaces, development of chemical and biological sensors, insulating layers in microelectronic circuits and fabrication of Nano devices just to name a few.
Various examples of protein SAM production methods are presented next:
1. Physical adsorption:
This approach is based on adsorption of proteins on such solid surfaces like carbon electrode, metal oxide or silica oxide. The adsorbed proteins constitute a self-assemble monolayer (SAM) with random orientations. Orientation control may be improved by modification of the protein and surface. This is demonstrated by Fig. 8.5(a).
2. Inclusion in polyelectrolytes or conducting polymers:
Polyelectrolytes or conducting polymers could provide a matrix in which the proteins are trapped and attached, or adsorbed, to the surface. This is demonstrated by Fig. 8.5(b).
3. Inclusion in SAM:
By using thiolated hydrocarbon chains, it is possible to produce a membrane-like monolayer on a noble metal, through which proteins can be located with non-specific orientations. Utilization of chains with different lengths (depressions and holes) would result in a SAM with definite topography that can give a specific orientation to the proteins. This is demonstrated by Fig. 8.5(c).
4. Non-oriented attachment to SAM:
In this approach the chains which form self-assemble monolayer (SAM) possess a functional group at their ends and react non-specifically with different parts of a protein. So, the orientation is random. This is demonstrated by Fig. 8.5(d).
5. Oriented attachment to SAM:
The principle is like the previous approach but the functional group here interacts specifically just with one domain or part of a given protein and hence a definite orientation is obtained. The structure of proteins can be chemically or genetically modified for this purpose. This is demonstrated by Fig 8.5(e).
6. Direct site-specific attachment to gold:
This is achieved by attachment of a unique cystine to gold surface. In this case the orientation can be completely controlled. This is demonstrated by Fig. 8.5(f).
A-5. Strain Directed self-assembly:
Strain directed self-assembly is applicable for fabrication and interconnection of wires and switches. In this approach a lithographically defined surface is impregnated with a strain of a compositionally controlled precipitation agent. It is possible to introduce a functional group to the substrate, which would couple with the surface functionality. This method can be used, for example, for semiconductor construction where it is necessary to immobilize system components on a solid support in order to gain total control over the progress and completion of the self-assembly process.
A-6. DNA Directed self-assembly:
DNA can be used, both, for site-selective immobilization and as a linker and thus provides a scaffold for nanostructure assembling. Nucleic acid-protein conjugates synthesis and utilization of specific interactions between two complementary DNA strands, antigen-anti- body and Bio-STV can bring about powerful tools to direct the mode of Nano-structure modules attachment.
Recent improvement in exerting genetic engineering techniques to the immobilized DNA sequences on a gold surface—like ligation, PCR (Polymerase Chain Reaction), and restriction digestion- provide even more control over self-assembly process. Fig. 8.6 illustrates utilization of DNA- STV conjugates in DNA directed assembly to construct nanostructures. Fig. 8.7 represents usage of biotin-streptavidin system in DNA- directed self-assembly.
Such a method can be applied to inorganic Nano crystal molecules. DNA can be also employed for template synthesis and its example is silver nanowire construction using DNA backbone. DNA has also been utilized to construct one-two- or three-dimensional frameworks.
It should be noted that DNA oligomer- tagged Nano modules could be attached together with favourite region-selectivity and in a controlled way. Detaching of an assembled nanostructure from its solid support leads to its folding and formation of an eventual conformation, the shape of which depends on the environmental conditions.
A-7. Self-assembly on silicon surfaces:
Employing silicon surfaces for self-assembly is of more importance in the micro/Nano electronics than other fields. Molecules and metals can be selectively deposited on a silicon template via such a method. This resembles patterning techniques to some extent (silicon based microlithography). Some other substances have been proposed for use in templated self-assembly like S-layer (single layer), proteins, diatom frustules, etc.
B. Self-assembly in Fluid Media:
Examples of self-assembly in a liquid, or in a dense-gas (supercritical fluid) phase, medium are:
(i) Self-assembly of polymerizable amphiphiles, polymer and lipid utilization to form ordered aggregates. Such ordered aggregates can be in the forms of monolayers, bilayers, micelles, micelle-coacervates or nanoparticles.
(ii) Fluidic self-assembly using a template surface and employing liquid stream to select appropriate components, is another approach. Fig. 8.8 illustrates the general procedure in fluidic self-assembly.
(iii) Dynamic combinatorial libraries (DCLs) which have an outstanding position in the combinatorial chemistry are the third approach.
(iv) Design of molecular cages through host-guest chemistry is yet another approach of fluidic self-assembly.
Conclusions and Discussion:
In this article, the factors responsible for, and categories of, self-assembly are presented and various examples of successful self-assemblies are introduced. Based on the material presented above we may conclude that self-assembly is a method of integration in which the components spontaneously assemble, typically by bouncing around in a solution or gas phase until equilibrium is reached (entropy is maximized and free energy is minimized) and a stable structure is generated.
Self-organization (which is the same as self-assembly) is the basic process (or driving force) that led up to the evolution of the biological world from inanimate matter. Understanding, inducing, and directing self- assembly is key to unravelling the progressive emergence of bottom-up nanotechnology. By understanding self-assembly we will be able to learn the role of various intermolecular interaction forces which govern a given self- assembly.
In order to induce and direct a desired self-assembly, it is also necessary, to be able to model and predict what happens under a certain process condition to an assembly of molecules and what directions they take, if any, to self-assemble.
While self-organization is crucial to the assembly of bio-molecular nanotechnology, it is also a promising method for assembling non-biological atoms and molecules, and especially molecular building blocks, towards creating materials with preferred behaviour and properties as well as precise devices. It should be emphasized that self-assembly is by no means limited to molecules or the Nano-scale. It can be carried out on just about any scale, making it a powerful bottom-up assembly method.