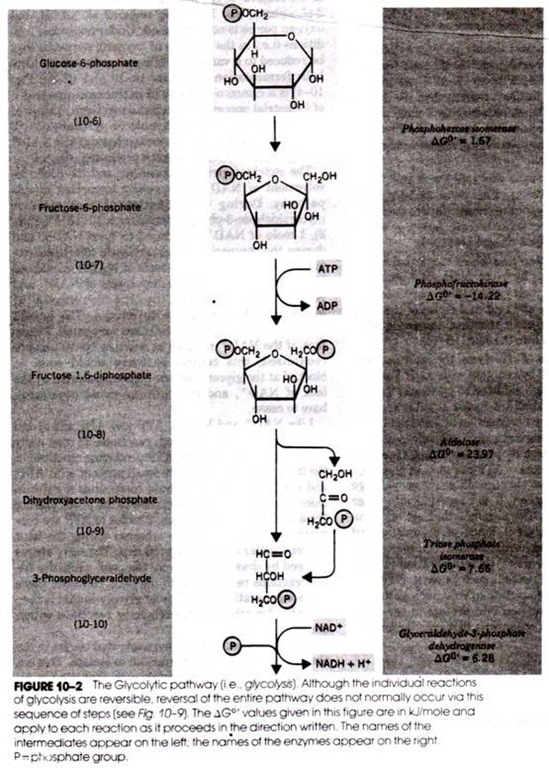
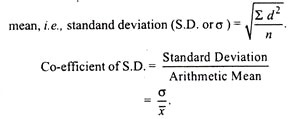
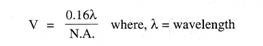Read this article to learn about the process of replication, recombination and repair of DNA.
Deoxyribonucleic acid (DNA) is a macromolecule that carries genetic information from generation to generation. It is responsible to preserve the identity of the species over millions of years. DNA may be regarded as a reserve bank of genetic information or a memory bank.
A single mammalian fetal cell contains only a few picograms (10-12 g) of DNA. It is surprising that this little quantity of DNA stores information that will determine the differentiation and every function of an adult animal.
Why did DNA evolve as genetic material?
RNA molecules, in principle, can perform the cellular functions that are carried out by DNA. In fact, many viruses contain RNA as the genetic material. Chemically, DNA is more stable than RNA. Hence, during the course of evolution, DNA is preferred as a more suitable molecule for long- term repository of genetic information.
The central dogma of life:
The biological information flows from DNA to RNA and from there to proteins. This is the central dogma of life (Fig. 3.1). It is ultimately the DNA that controls every function of the cell through protein synthesis.
As the carrier of genetic information, DNA in a cell must be duplicated (replicated), maintained and passed down accurately to the daughter cells. Three distinct processes are designed for this purpose. The ‘three Rs’ of DNA-replication, recombination, and repair. There are certain common features between the three Rs.
I. They act on the same substrate (DNA).
II. They are primarily concerned with the making and breaking of phosphodiester bonds (the backbone of DNA structure).
III. Enzymes used in the three processes are mostly similar/ comparable.
Replication of DNA:
DNA is the genetic material. When the cell divides, the daughter cells receive an identical copy of genetic information from the parent cell. Replication is a process in which DNA copies itself to produce identical daughter molecules of DNA Replication is carried out with high fidelity which is essential for the survival of the species.
Synthesis of a new DNA molecule is a complex process involving a series of steps. The salient features of replication in prokaryotes are described first. This is followed by some recent information on the eukaryotic replication.
Replication in Prokaryotes:
Replication is semiconservative:
The parent DNA has two strands complementary to each other. Both the strands undergo simultaneous replication to produce two daughter molecules. Each one of the newly synthesized DNA has one-half of the parental DNA (one strand from original) and one-half of new DNA (Fig. 3.2).
This type of replication is known as semiconservative since half of the original DNA is conserved in the daughter DNA. The first experimental evidence for the semiconservative DNA replication was provided by Meselson and Stahl (1958).
Initiation of replication:
The initiation of DNA synthesis occurs at a site called origin of replication. In case of prokaryotes, there is a single site whereas in eukaryotes, there are multiple sites of origin. These sites mostly consist of a short sequence of A-T base pairs. A specific protein called dna A (20-50 monomers) binds with the site of origin for replication. This causes the double-stranded DNA to separate.
Replication bubbles:
The two complementary strands of DNA separate at the site of replication to form a bubble. Multiple replication bubbles are formed in eukaryotic DNA molecules, which is essential for a rapid replication process (Fig. 3.3).
RNA primer:
For the synthesis of new DNA, a short fragment of RNA (about 5-50 nucleotides, variable with species) is required as a primer. This primer is synthesized on the DNA template by a specific RNA polymerase called primase. A constant synthesis and supply of RNA primers should occur on the lagging strand of DNA. This is in contrast to the leading strand which has almost a single RNA primer.
DNA synthesis is semi discontinuous and bidirectional:
The replication of DNA occurs in 5′ to 3′ directions, simultaneously, on both the strands of DNA. On one strand, the leading (continuous or forward) strand—the DNA synthesis is continuous. On the other strand, the lagging (discontinuous or retrograde) strand—the synthesis of DNA is discontinuous. Short pieces of DNA (15-250 nucleotides) are produced on the lagging strand. In the replication bubble, the DNA synthesis occurs in both the directions (bidirectional) from the point of origin.
Replication fork and DNA synthesis:
The separation of the two strands of parent DNA results in the formation of a replication fork. The active synthesis of DNA occurs in this region. The replication fork moves along the parent DNA as the daughter DNA molecules are synthesized.
DNA helicases:
These enzymes bind to both the DNA strands at the replication fork. Helicases move along the DNA helix and separate the strands. Their function is comparable with a zip opener. Helicases are dependent on ATP for energy supply.
Single-stranded DNA binding (SSB) proteins:
These are also known as DNA helix-destabilizing proteins. They possess no enzyme activity. SSB proteins bind only to single-stranded DNA (separated by helicases), keep the two strands separate and provide the template for new DNA synthesis. It is believed that SSB proteins also protect the single-stranded DNA degradation by nucleases.
DNA synthesis catalysed by DNA polymerase III:
The synthesis of a new DNA strand, catalysed by DNA polymerase III, occurs in 5’—>3′ direction. This is antiparallel to the parent template DNA strand. The presence of all the four de-oxyribo-nucleoside triphosphates (dATP, dGTP, dCTP and dTTP) is an essential prerequisite for replication to take place.
The synthesis of two new DNA strands, simultaneously, takes place in the opposite direction—one is in a direction (5’→3′) towards the replication fork which is continuous, the other in a direction (5’→3′) away from the replication fork which is discontinuous (Fig. 3.4).
The incoming deoxyribonucleotides are added one after another, to 3′ end of the growing DNA chain (Fig. 3.5). A molecule of pyrophosphate (PPi) is removed with the addition of each nucleotide. The template DNA strand (the parent) determines the base sequence of the newly synthesized complementary DNA.
Polarity problem:
The DNA strand (leading strand) with its 3′-end (3′-OH) oriented towards the fork can be elongated by sequential addition of new nucleotides. The other DNA strand (lagging strand) with 5′-end presents some problem, as there is no DNA polymerase enzyme (in any organism) that can catalyse the addition of nucleotides to the 5′ end (i.e. 3’→5′ direction) of the growing chain. This problem however is solved by synthesizing this strand as a series of small fragments. These pieces are made in the normal 5’→3′ direction, and later joined together.
Okazaki pieces:
The small fragments of the discontinuously synthesized DNA are called Okazaki pieces. These are produced on the lagging strand of the parent DNA. Okazaki pieces are later joined to form a continuous strand of DNA. DNA polymerase I and DNA ligase are responsible for this process (details given later).
Proof-reading function of DNA polymerase III:
Fidelity of replication is the most important for the very existence of an organism. Besides its 5’→3′ directed catalytic function, DNA polymerase III also has a proof-reading activity. It checks the incoming nucleotides and allows only the correctly matched bases (i.e. complementary bases) to be added to the growing DNA strand. Further, DNA polymerase edits its mistakes (if any) and removes the wrongly placed nucleotide bases.
Replacement of RNA primer by DNA:
The synthesis of new DNA strand continues till it is in close proximity to RNA primer. Now the DNA polymerase I comes into picture. It removes the RNA primer and takes its position. DNA polymerase I catalyses the synthesis (5’→3′ direction) of a fragment of DNA that replaces RNA primer (Fig. 3.6).
The enzyme DNA ligase catalyses the formation of a phosphodiester linkage between the DNA synthesized by DNA polymerase III and the small fragments of DNA produced by DNA polymerase I. This process—nick sealing-requires energy, provided by the breakdown of ATP to AMP and PPi. Another enzyme—DNA polymerase II—has been isolated. It participates in the DNA repair process.
Supercoils and DNA topoisomerases:
As the double helix of DNA separates from one side and replication proceeds, supercoils are formed at the other side. The formation of supercoils can be better understood by comparing DNA helix with two twisted ropes tied at one end. Hold the ropes at the tied end in a fixed position. And let your friend pull the ropes apart from the other side. The formation of supercoils is clearly observed.
The problem of supercoils that comes in the way of DNA replication is solved by a group of enzymes called DNA topoisomerases. Type I DNA topoisomerase cuts the single DNA strand (nuclease activity) to overcome the problem of supercoils and then reseals the strand (ligase activity). Type II DNA topoisomerase (also known as DNA gyrase) cuts both strands and reseals them to overcome the problem of supercoils.
Replication in Eukaryotes:
Replication of DNA in eukaryotes closely resembles that of prokaryotes. Certain differences, however, exist. Multiple origins of replication is a characteristic feature of eukaryotic cell. Further, at least five distinct DNA polymerases are known in eukaryotes.
Greek letters are used to number these enzymes:
1. DNA polymerase α is responsible for the synthesis of RNA primer for both the leading and lagging strands of DNA.
2. DNA polymerase β is involved in the repair of DNA. Its function is comparable with DNA polymerase I found in prokaryotes.
3. DNA polymerase γ this enzyme participates in the replication of mitochondrial DNA.
4. DNA polymerase δ is responsible for the replication on the leading strand of DNA. It also possesses proof-reading activity.
5. DNA polymerase ε is involved in DNA synthesis on the lagging strand and proof-reading function.
The differences in the DNA replication between bacteria and human cells, attributed to the enzymes, are successfully used in antibacterial therapy to target pathogen (bacterial) replication and spare the host (human) cells.
Process of Replication in Eukaryotes:
The replication on the leading (continuous) stand of DNA is rather simple, involving DNA polymerase δ and a sliding clamp called proliferating cell nuclear antigen (PCNA). PCNA is so named as it was first detected as an antigen in the nuclei of replicating cells. PCNA forms a ring around DNA to which DNA polymerase S binds. Formation of this ring also requires another factor namely replication factor C (RFC).
The replication on the lagging (discontinuous) strand in eukaryotes is more complex when compared to prokaryotes or even the leading strand of eukaryotes. This is depicted in Fig. 3.7, and briefly described hereunder.
The parental strands of DNA are separated by the enzyme helicase. A single-stranded DNA binding protein called replication protein A (RPA) binds to the exposed single-stranded template. This strand has been opened up by the replication fork (a previously formed Okazaki fragment with an RNA primer is also shown in Fig. 3.4).
The enzyme primase forms a complex with DNA polymerase α which initiates the synthesis of Okazaki fragments. The primase activity of pol α-primase complex is capable of producing 10-bp RNA primer. The enzyme activity is then switched from primase to DNA polymerase α which elongates the primer by the addition of 20-30 deoxyribonucleotides. Thus, by the action of pol α-primase complex, short stretch of DNA attached to RNA is formed. And now the complex dissociates from the DNA.
The next step is the binding of replication factor C (RFC) to the elongated primer (short RNA-DNA). RFC serves as a clamp loader and catalyses the assembly of proliferating cell nuclear antigen (PCNA) molecules. The DNA polymerase 8 binds to the sliding clamp and elongates the Okazaki fragment to a final length of about 150-200 bp. By this elongation, the replication complex approaches the RNA primer of the previous Okazaki fragment.
The RNA primer removal is carried out by a pair of enzymes namely RNase H and flap endonuclease I (FEND. This gap created by RNA removal is filled by continued elongation of the new Okazaki fragment (carried out by polymerase 8, described above). The small nick that remains is finally sealed by DNA ligase.
Eukaryotic DNA is tightly bound to histones (basic proteins) to form nucleosomes which, in turn, organize into chromosomes. During the course of replication, the chromosomes are relaxed and the nucleosomes get loosened. The DNA strands separate for replication, and the parental histones associate with one of the parental strands.
As the synthesis of new DNA strand proceeds, histones are also produced simultaneously, on the parent strand. At the end of replication, of the two daughters chromosomal DNAs formed, one contains the parental histones while the other has the newly synthesized histones.
Inhibitors of DNA Replication:
Bacteria contain a specific type II topoisomerase namely gyrase. This enzyme cuts and reseals the circular DNA (of bacteria), and thus overcomes the problem of supercoils. Bacterial gyrase is inhibited by the antibiotics ciprofloxacin, novobiocin and nalidixic acid. These are widely used as antibacterial agents since they can effectively block the replication of DNA and multiplication of cells. These antibacterial agents have almost no effect on human enzymes.
Certain compounds that inhibit human topoisomerases are used as anticancer agents e.g. adriamycin, etoposide, doxorubicin. The nucleotide analogs that inhibit DNA replication are also used as anticancer drugs e.g. 6-mercaptopurine, 5-fluorouracil.
Cell Cycle and DNA Replication:
The cell cycle consists of four distinct phases in higher organisms—mitotic, G1, S and G2 phases (Fig. 3.8). When the cell is not growing, it exists in a dormant or un-dividing phase (G0). G1 phase is characterized by active protein synthesis.
Replication of DNA occurs only once in S-phase and the chromosomes get doubled i.e. diploid genome gets converted into tetraploid. The entire process of new DNA synthesis takes place in about 8-10 hours and a large number of DNA polymerases (500-1,000) are simultaneously involved in this process. It is believed that methylation of DNA serves as a marker to inhibit replication. The G2 phase is characterized by enlargement of cytoplasm and this is followed by the actual cell division that occurs in the mitotic phase.
Cyclins and cell cycle:
Cyclins are a group of proteins that are closely associated with the transition of one phase of cell cycle to another, hence they are so named. The most important cyclins are cyclin A, B, D and E. The concentrations of cyclins increase or decrease during the course of cell cycle. These cyclins act on cyclin-dependent kinases (CDKs) that phosphorylate certain substances essential for the transition of one cycle to another.
Cyclins and cyclin-dependent kinases (CDK1, CDK2, CDK4, and CDK6) are intimately connected with the progression of cell cycle. For instance, cyclin D levels rise in late G1 phase which activate CDK4 and CDK6. This results in the assembly of nuclear proteins in a complex form in late G1 phase.
Cell cycle check points:
As depicted in Fig. 3.8, there occurs a continuous monitoring of the cell cycle with respect to DNA replication, chromosome segregation and integrity. If any damage to DNA is detected either in G1 or G2 phase of the cycle, or if there is a formation of defective spindle (i.e. incomplete chromosomal segregation), the cell cycle will not progress until appropriately corrected. If it is not possible to repair the damage done, the cells undergo apoptosis (programmed cell death).
Cancer and cell cycle:
Cancer represents an excessive division of cells. In cancer, a large quantity of cells are in mitosis and most of them in S-phase. Majority of the drugs used for cancer therapy are designed to block DNA replication or inhibit the enzymes that participate in replication (directly or indirectly). Methotrexate (inhibits dihydrofolate reductase) and 5-fluorouracil (inhibits thymidylate synthase) block nucleotide synthesis. In recent years, topoisomerase inhibitors are being used. They block the unwinding of parental DNA strands and prevent replication.
Telomeres and Telomerase:
There are certain difficulties in the replication of linear DNAs (or chromosomes) of eukaryotic cells. The leading strand of DNA can be completely synthesized to the very end of its template. This is not possible with the lagging strand, since the removal of the primer RNA leaves a small gap which cannot be filled (Fig. 3.9A).
Consequently, the daughter chromosomes will have shortened DNA molecules. This becomes significant after several cell cycles involving replication of chromosomes. The result is that over a period of time, the chromosomes may lose certain essential genes and the cell dies. This is however, avoided to a large extent.
Telomeres are the special structures that prevent the continuous loss of DNA at the end of the chromosomes during the course of replication. Thus, they protect the ends of the chromosomes, and are also responsible to prevent the chromosomes from fusing with each other. Telomeres are many repeat sequences of six nucleotides present at the ends of eukaryotic chromosomes. Human telomeres contain thousands of repeat TTACGC sequences, which can be up to a length of 1500 bp.
Role of telomerase:
Telomeres are maintained by the enzyme telomerase, also called as telomere terminal transferase. Telomerase is an unusual enzyme as it is composed of both protein and RNA. In case of humans, the RNA component is 450 nucleotides in length, and at the 5′-terminal and it contains the sequence 5′-CUAACCCUAAC-3′.
It may be noted that the central region of this sequence is complementary to the telomere repeat sequence 5′-TTAGGG-3′. The telomerase RNA sequence can be used as a template for extension of telomeres (Fig. 3.9B).
The telomerase RNA base pairs to the end of the DNA molecule with telomeres and extends to a small distance. Then translocation of telomerase occurs and a fresh extension of DNA takes place. This process of DNA synthesis and translocation is repeated several times until the chromosome gets sufficiently extended. The extension process gets completed through the participation of DNA polymerase and primase complex and sealing of the new DNA formed.
It may be noted here that as such the telomeres do not encode proteins. Hence, when extended by telomerase, they need not have to remain the same length, and some shortening will not pose any problem. During the course of repeated cell cycles, there occurs progressive shortening of telomeres, and this has to be prevented, which is appropriately carried out by telomerase.
Telomere in Senescence and Cancer:
Evidence is now forthcoming that telomerase is not active in all the mammalian cells. This is mainly because cells that have undergone differentiation no longer divide or divide only to a limited extent. Telomerase is highly active in the early embryo, and after birth it is active in the reproductive and stem cells.
Stem cells divide continuously throughout the lifetime of an organism to produce new cells. These cells in turn are responsible to tissues and organs in the functional state e.g. hematopoietic stem cells of bone marrow.
Many biologists link the process of telomere shortening with cell senescence (i.e. cell death). This is mainly based on the observations made in the in vitro mammalian cell cultures. However, some researchers question this relation between telomere shortening and senescence.
Cancerous cells are able to divide continuously. There is a strong evidence to suggest that the absence of senescence in cancer cells is linked to the activation of the enzyme telomerase. Thus, telomere length is maintained throughout multiple cell divisions. It is however, not clear whether telomerase activation is a cause or an effect of cancer.
There is however, evidence to suggest that telomerase activation is in fact the cause of certain cancers e.g. dyskeratosis congenita due to a mutation in the gene responsible for the RNA component of telomerase. The enzyme telomerase is an attractive target for cancer chemotherapy. The drugs have been designed to inactivate telomerase, and consequently induce senescence in the cancer cells. This in turn prevents the rapid cell proliferation.
Recombination:
Recombination basically involves the exchange of genetic information. There are mainly two types of recombination’s.
1. Homologous recombination:
This is also called as general recombination, and occurs between identical or nearly identical chromosomes (DNA sequences). The best example is the recombination between the paternal and maternal chromosomal pairs (Fig. 3.10).
2. Non-homologous recombination:
This is regarded as illegitimate recombination and does not require any special homologous sequences. Transposition is a good example of non-homologous recombination. Random integration of outside genes into mammalian chromosomes is another example.
Homologous Recombination:
It is a known fact that the chromosomes are not passed on intact from generation to generation. Instead, they are inherited from both the parents. This is possible due to homologous recombination. Three models have been put forth to explain homologous recombination’s.
i. Holliday model
ii. Meselson-Radding model
iii. Double-strand break model.
Holliday model:
Holliday model (proposed by Holliday in 1964) is the simplest among the homologous recombination models. It is depicted in Fig. 3.11
The two homologous chromosomes come closer, get properly aligned, and form single-strand breaks. This results in two aligned DNA duplexes. Now the strands of each duplex partly unwind and invade in the opposite direction to form a two strands cross between the DNA molecules.
There occurs simultaneous unwinding and rewinding of the duplexes in such a way that there is no net change in the amount of base pairing, but the position of crossover moves. This phenomenon referred to as branch migration, results in the formation of heteroduplex DNA.
The enzyme DNA ligase seals the nick. The two DNA duplexes (4 strands of DNA), joined by a single crossover point can rotate to create a four-standard Holliday junction. Now the DNA molecules are subjected to symmetrical cuts in either of the two directions, and the cut ends are resealed by ligase.
The DNA exchange is determined by the direction of the cuts, which could be horizontal or vertical. If the cross strands are cut horizontally (cut 1), the flanking genes (or markers, i.e. AB/ab) remain intact, and no recombination occurs. On the other hand, if the parental strands are cut vertically (cut 2), the flanking genes get exchanged (i.e. Ab/aB) due to recombination.
Non-Homologous Recombination:
The recombination process without any special homologous sequences of DNA is regarded as non-homologous recombination.
Transposition:
Transposition primarily involves the movement of specific pieces of DNA in the genome. The mobile segments of DNA are called transposons or transposable elements. They were first discovered by Barbara McClintock (in 1950) in maize, and their significance was ignored for about two decades by other workers.
Transposons are mobile and can move almost to any place in the target chromosome. There are two modes of transposition. One that involves an RNA intermediate and the other which does not involve RNA intermediate.
Retro transposition:
Transposition involving RNA intermediate represents retro transposition (Fig. 3.12). By the normal process of transcription, a copy of RNA formed from a transposon (also called as retro transposon). Then by the enzyme reverse transcriptase, DNA is copied from the RNA.
The newly formed DNA which is a copy of the transposon gets integrated into the genome. This integration may occur randomly on the same chromosome or, on a different chromosome. As a result of the retro transposition, there are now two copies of the transposon, at different points on the genome.
DNA transposition:
Some transposons are capable of direct transposition of DNA to DNA. This may occur either by replicative transposition or conservative transposition (Fig. 3.13). Both the mechanisms require enzymes that are mostly coded by the genes within the transposons.
In the replicative transposition, a direct interaction occurs between the donor transposon and the target site to result in copying of the donor element.
In case of conservative transposition, the transposon is excised and reintegrated at a new site.
DNA transposition is less common than retro transposition in case of eukaryotes. However, in case of prokaryotes, DNA transposons are more important than RNA transposons.
Significance of transposition:
It is now widely accepted that a large fraction of the human genome has resulted due to the accumulation of transposons. Short interspersed elements (SINEs) are repeats of DNA sequences which are present in about 500,000 copies per haploid human genome e.g. Alu sequences.
Long interspersed elements (LINEs) are also repeated DNA sequences and are present in about 50,000 copies in the human genome e.g. L1 elements. Some of the diseases caused by mutations are due to insertion of transoms into a genes.
Damage and Repair of DNA:
Being the carrier of genetic information, the cellular DNA must be replicated (duplicated), maintained, and passed down to the daughter cells accurately. In general, the accuracy of replication is extremely high. However, there do occur replication errors. It is estimated that approximately one error is introduced per billion base pairs during each cycle of replication. The cells do possess the capability to repair damages done to DNA to a large extent.
Consequences of DNA Damage:
Despite an efficient repair system for the damaged DNA, replication errors do accumulate that ultimately result in mutations. The human body possesses 1014 nucleated cells, each with 3 × 109 base pairs of DNA. It is estimated that about 1016 cell divisions occur in a lifetime. If 10-10 mutations per base pair per cell generation escape repair, this results in about one mutation per 106 base pairs in genome.
Besides the possible errors in replication, the DNA is constantly subjected to attack by both physical and chemical agents. These include radiation, free radicals, and chemicals etc., which also result in mutations.
It is fortunate that a great majority of the mutations probably occur in the DNA that does not encode proteins, and consequently will not have any serious impact on the organism. This is not, however, all the time true, since mutations do occur in the coding regions of DNA also. There are situations in which the change in a single base pair in the human genome can cause a serious disease e.g. sickle-cell anemia.
Types of DNA Damages:
The damage done to DNA by physical, chemical and environmental agents may be broadly classified into four categories with different types (Table 3.1).
The DNA damage may occur due to single-base alterations (e.g. depurination, deamination), two- base alterations (e.g. pyrimidine diamer) chain breaks (e.g. ionizing radiation) and cross-linkages (e.g. between bases). Some selected DNA damages are briefly described. The occurrence of spontaneous deamination bases in aqueous solution at 37°C is well known. Cytosine gets deaminated to form uracil while adenine forms hypoxanthine.
Spontaneous depurination, due to cleavage of glycosyl bonds (that connect purines to the backbone) also occurs. It is estimated that 2000-10,000 purines may be lost per mammalian cell in 24 hours. The depurinated sites are called as abasic sites. Originally, they were detected in purines, and called apurinic sites (AP sites) which represent lack of purine. Now, the term AP sites is generally used to represent any base lacking in DNA.
The production of reactive oxygen species is often associated with alteration of bases e.g. formation of 8-hydroxy guanine. Free radical formation and oxidative damage to DNA increases with advancement of age. Ultraviolet radiations result in the formation of covalent links between adjacent pyrimidine’s along the DNA strand to form pyrimidine dimers. DNA chain breaks can be caused by ionizing radiations (e.g. X-rays).
Mutations:
The genetic macromolecule DNA is highly stable with regard to its base composition and sequence. However, DNA is not totally exempt from gradual change. Mutation refers to a change in the DNA structure of a gene. The substances (chemicals) which can induce mutations are collectively known as mutagens. The changes that occur in DNA on mutation are reflected in replication, transcription and translation.
Types of mutations:
Mutations are mainly of two major types—point mutations, frame shift mutations (Fig. 3.14).
1. Point mutations:
The replacement of one base pair by another results in point mutation. They are of two sub-types.
(a) Transitions:
In this case, a purine (or a pyrimidine) is replaced by another.
(b) Trans-versions:
These are characterized by replacement of a purine by a pyrimidine or vice versa.
2. Frame-shift mutations:
These occur when one or more base pairs are inserted in or deleted from the DNA, respectively, causing insertion or deletion mutations.
Consequences of point mutations:
The change in a single base sequence in point mutation may cause one of the following (Fig. 3.15).
1. Silent mutation:
The codon (of mRNA) containing the changed base may code for the same amino acid. For instance, UCA codes for serine and change in the third base (UCU) still codes for serine. This is due to degeneracy of the genetic code. Therefore, there are no detectable effects in silent mutation.
2. Missense mutation:
In this case, the changed base may code for a different amino acid. For example, UCA codes for serine while ACA codes for threonine. The mistaken (or missense) amino acid may be acceptable, partially acceptable or unacceptable with regard to the function of protein molecule. Sickle-cell anemia is a classical example of missense mutation.
3. Nonsense mutation:
Sometimes, the codon with the altered base may become a termination (or nonsense) codon. For instance, change in the second base of serine codon (UCA) may result in UAA. The altered codon acts as a stop signal and causes termination of protein synthesis, at that point.
Consequences of frame shift mutations:
The insertion or deletion of a base in a gene results in an altered reading frame of the mRNA (hence the name frame shift). The machinery of mRNA (containing codons) does not recognize that a base was missing or a new base was added. Since there are no punctuations in the reading of codons, translation continues. The result is that the protein synthesized will have several altered amino acids and/or prematurely terminated protein.
Mutations and cancer:
Mutations are permanent alterations in DNA structure, which have been implicated in the etiopathogenesis of cancer.
Repair of DNA:
As already stated, damage to DNA caused by replication errors or mutations may have serious consequences. The cell possesses an inbuilt system to repair the damaged DNA. This may be achieved by four distinct mechanisms (Table 3.2).
1. Base excision-repair
2. Nucleotide excision-repair
3. Mismatch repair
4. Double-strand break repair.
1. Base excision-repair:
The bases cytosine, adenine and guanine can undergo spontaneous depurination to respectively form uracil, hypoxanthine and xanthine. These altered bases do not exist in the normal DNA, and therefore need to be removed. This is carried out by base excision repair (Fig. 3.16).
A defective DNA in which cytosine is deaminated to uracil is acted upon by the enzyme uracil DNA glycosylase. This results in the removal of the defective base uracil. An endonuclease cuts the backbone of DNA strand near the defect and removes a few bases. The gap so created is filled up by the action of repair DNA polymerase and DNA ligase.
2. Nucleotide excision-repair:
The DNA damage due to ultraviolet light, ionizing radiation and other environmental factors often results in the modification of certain bases, strand breaks, cross-linkages etc. Nucleotide excision-repair is ideally suited for such large-scale defects in DNA. After the identification of the defective piece of the DNA, the DNA double helix is unwound to expose the damaged part.
An excision nuclease (exinuclease) cuts the DNA on either side (upstream and downstream) of the damaged DNA. This defective piece is degraded. The gap created by the nucleotide excision is filled up by DNA polymerase which gets ligated by DNA ligase (Fig. 3.17).
Xeroderma pigmentosum (XP) is a rare autosomal recessive disease. The affected patients are photosensitive and susceptible to skin cancers. It is now recognized that XP is due to a defect in the nucleotide excision repair of the damaged DNA.
Mismatch repair:
Despite high accuracy in replication, defects do occur when the DNA is copied. For instance, cytosine (instead of thymine) could be incorporated opposite to adenine. Mismatch repair corrects a single mismatch base pair e.g. C to A, instead of T to A.
The template strand of the DNA exists in a methylated form, while the newly synthesized strand is not methylated. This difference allows the recognition of the new strands. The enzyme GATC endonuclease cuts the strand at an adjacent methylated GATC sequence (Fig. 3.18). This is followed by an exonuclease digestion of the defective strand, and thus its removal. A new DNA strand is now synthesized to replace the damaged one.
Hereditary non-polyposis colon cancer (HNPCC) is one of the most common inherited cancers. This cancer is now linked with faulty mismatch repair of defective DNA.
Double-strand break repair:
Double-strand breaks (DSBs) in DNA are dangerous. They result in genetic recombination which may lead to chromosomal translocation, broken chromosomes, and finally cell death. DSBs can be repaired by homologous recombination or non-homologous end joining. Homologous recombination occurs in yeasts while in mammals, non-homologous and joining dominates.
Defects in DNA Repair and Cancer:
Cancer develops when certain genes that regulate normal cell division fail or are altered. Defects in the genes encoding proteins involved in nucleotide-excision repair, mismatch repair and re-combinational repair are linked to human cancers. For instance, HNPCC is due to a defect in mismatch repair.