In this article we will learn about:- 1. Origin of Inversions 2. Types of Inversion 3. Effects 4. Uses.
Origin of Inversions:
Inversion was first discovered by Sturtevant in 1921 in Drosophila. Later it was detected in a wide variety of plant and animal species. Inversions occur spontaneously, and can be induced artificially. In many organisms, inversions are found in the natural populations.
Inversions have been studied extensively in different Drosophila species, such as, D. pseudoobscura, D. persimilis and D. willistoni. They have been reported in several plant species like maize, barley, broad bean, Tradescantia etc. Inversions have been induced in several plant species through mutagenic treatments.
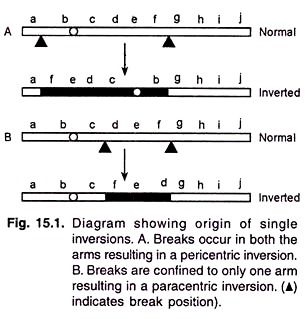
Inversion involves at least two breaks in a chromosome; the broken segment rotates 180° and is then reunited at the two break points.
Types of Inversion:
Based on the number of inverted segments within a chromosome and the location of the inversion points with respect of each other, the inversion may be broadly classified into two types:
(1) Single inversion and
(2) Complex inversion.
1. Single inversion:
In this case, only one segment of the chromosome is inverted. There are two types of single inversion:
(i) Pericentric inversion:
The inverted segment includes the centromere, i.e., the two breakpoints are located in different arms of the chromosome (Fig. 15.1A).
(ii) Paracentric inversion:
The inversion is confined to a single arm of the chromosome, i.e., both the inversion breakpoints are located in the same arm (Fig. 15.1B).
2. Complex inversion:
Occurrence of more than one inversion in a chromosome is called complex inversion. Based on the mutual relationship of the inverted regions, it may be grouped into the following five types.
(i) Independent inversions:
Inversions occur in different regions of the chromosome and they are separated from one another by un-inverted (normal) segment (Fig. 15.2A).
(ii) Direct tandem inversions:
There are two or more inverted segments which are directly adjacent to each other, i.e., the inverted regions are not separated by normal regions (Fig. 15.2B).
(iii) Reversed tandem inversions:
The two inverted segments are adjacent to each other but their positions are mutually interchanged i.e., g. the first segment lies in place of the second and vice-versa (Fig. 15.2C).
(iv) Included inversions:
One inversion is confined within another inversion, i.e., a segment within an inverted segment is inverted again; as a result, the second inverted segment possesses the normal gene sequence for the concerned segment (Fig. 15.2D).
(v) Overlapping inversions:
Such inversions have a common segment, i.e., a part of an inverted chromosome segment is inverted again together with an adjacent segment which was not included in the first inverted segment (Fig. 15.2E).
Effects of Inversion:
1. Effect on Fertility:
Fertility of inversion heterozygotes is reduced due to the production of unbalanced gametes which carry the deficiency-duplication chromatids obtained from crossing over within the inversion loop. The effect on fertility varies according to the type of inversion and the organism carrying it (Table 15.4).
2. Recessive mutations:
Damage to the DNA at the breakpoints may sometimes result in a recessive mutation. Some of the mutations may be recessive lethals; such mutations are also lethal in hemizygous condition.
3. Position effect:
Due to inversion euchromatic segment of a chromosome may become located in the vicinity of a heterochromatic region. In such a condition, the euchromatic region may become hetero-chromatinized; this would cause the suppression of gene activity i.e., suppression of transcription of the euchromatic segment. Heterochromatinization is variable as a consequence of which variegated type (V-type) position effects are produced.
For example, in Drosophila, the gene for red eye (w+), if joined to the heterochromatic region due to an inversion, expresses like the recessive allele for white eye (w). In heterozygotes (w+lw), a variegation of red and white spots in the eye are observed. But when the inversion is reverted, red eye colour is regularly produced.
4. Effect of inversion on crossing over in the non-inverted region of the same chromosome and in other chromosomes of the complement:
There are instances where heterozygous inversion in a chromosome pair affects the crossing over in other non-inverted regions of the same chromosome (intra-chromosomal effect) or in the non-homologous chromosomes (inter-chromosomal effect). Generally the heterozygous inversion in a chromosome causes an increase in crossing over in the non-homologous chromosome pairs of the same complement.
Thus the reduced crossing over in the inverted region seems to be compensated by an increased recombination in the remaining bivalents as if to maintain the optimal frequency of crossing over. This phenomenon is called “Schultz-Redfield effect” after the name of the scientists who discovered it in Drosophila.
5. Effect of inversion on the activity of nucleolar organizer region (NOR) of other chromosome:
Inversion in one chromosome influences the NOR activity in the other chromosomes. Viseras and Camacho in 1991 did not find decreased activity of NOR in L3 does chromosome due to the presence of a pericentric inversion in the smallest chromosome (S11 chromosome) of grasshopper Aiolopusstrepen.
NOR activity was significantly reduced from normal when the pericentric inversion was in homozygous condition (Table 15.5).
Uses of Inversion:
1. Inversion can be used to study the behaviour of chromosomes during meiosis, such as, chromosome pairing, cytological crossing over and formation of bridges and fragments and various configurations at different meiotic stages.
2. Paracentric inversion can be used to produce acentric fragments, so that behaviour and fate of these fragments can be studied.
3. Asymmetrical inversion breakpoints on the two sides of centromere in a pericentric inversion will change the karyotype of the chromosome. A metacentric chromosome may become a sub-metacentric or acrocentric depending on the break positions in the two arms (Fig. 15.5). Thus karyotype polymorphism may be generated.
4. A combination of cytological location of inversion breakpoints and genetic linkage can be used for physical location of genes in the chromosome.
5. Absence of crossing over within the inverted segment maintains the particular gene combination intact, i.e., it produces complete linkage for the concerned genes. Such inversion behaves like a dominant gene designated as crossover suppressor.
6. Crossover suppressors (inversions) can be used to study the specific problems in genetics. The famous “CIB” technique of Muller is an important example of this type. In “CIB”, C represents the crossover suppressor gene (inverted chromosome segment), 1 represents a recessive lethal in homozygous and hemizygous conditions, and B represents the ‘Bar-eye’ in Drosophila-, these genes are located in the X chromosome.
Muller in 1927 used this stock to detect the lethal mutations induced by X-rays.
7. Inversions can be used to study the behaviour of the broken ends of chromosomes. The broken ends may fuse to produce dicentric chromatid bridge.
8. Inversions have played an important role in evolution. Various kinds of complex inversions may produce different races. The phylogenetic relations of different species can be studied, especially for overlapping inversions.
9. The location of genes for desirable characters can be done using inversions. Their use in locating the favourable genes for quantitative characters carried by inbreds was suggested by Dobzhansky and Rhoades in 1938. In 1941, Sprague used pericentric inversion and in 1959 Chao used paracentric inversion stocks for such studied in maize inbred lines.
10. Inversions can be used to study the position effect.
11. Inversions may be utilized in combination with other chromosomal changes for genetic control of insects as suggested by Bhalla in 1970 for yellow fever mosquito (Aedesaegypti).